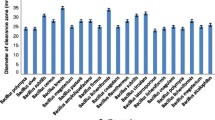
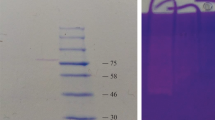
Abstract
Bacterial proteases have extensive applications in various fields of industrial microbiology. In this study, protease-producing organisms were screened on skimmed milk agar media using serial dilution. Through microbial biomass production, biochemical tests, protease-specific activity, and 16 s RNA gene sequencing, the isolates were identified as Bacillus subtilis and submitted to NCBI. The strain accession numbers were designated as A1 (MT903972), A2 (MT903996), A4 (MT904091), and A5 (MT904796). The strain A4 Bacillus subtilis showed highest protease-specific activity as 76,153.84 U/mg. A4 Bacillus subtilis was unaffected by Ca2+, Cu2+, Fe2+, Hg2+, Mg2+, Na, Fe2+, and Zn2+ but was inhibited by 80% by Mn2+ (5 mM). The protease activity was inhibited by up to 30% by iodoacetamide (5 mM). These findings confirm the enzyme to be a cysteine protease which was further confirmed by MALDI-TOF. The identified protease showed 71% sequence similarity with Bacillus subtilis cysteine protease. The crude cysteine protease significantly aided in fabric stain removal when added to a generic detergent. It also aided in the recovery of silver from used X-ray films and de-hairing of goat skin hides and showed decent application in meat tenderization. Thus, the isolated cysteine protease has high potential for industrial applications.






Similar content being viewed by others
Data availability
The data that support the findings of this study are available on request from the corresponding author, SB.
References
Paula S, Mona A, Carolina C et al (2015) A biotechnology perspective of fungal proteases. Braz J Microbiol 46(2):337–346. https://doi.org/10.1590/2FS1517-838246220140359
Barrett AJ, Rawlings ND, O’Brien EA (2001) The MEROPS database as a protease information system. J struct boil 134(2–3):95–102. https://doi.org/10.1006/jsbi.2000.4332
Asha B, Palaniswamy M (2018) Optimization of alkaline protease production by Bacillus cereus FT 1 isolated from soil. J Appl Pharm Sci 8(02):119–127. https://doi.org/10.7324/JAPS.2018.8219
Theron W, Divol B (2014) Microbial aspartic proteases: current and potential applications in industry. Appl Microbiol Biotechnol 98:8853–8868. https://doi.org/10.1007/s00253-014-6035-6
Satbir S, Bijender K (2017) Potential application spectrum of microbial proteases for clean and green industrial production. Energ. Ecol. Environ 2(6):370–386. https://doi.org/10.1007/s40974-017-0076-5#citeas
Yogesh P, Akshaya G, Shilpa G (2018) Production, partial purification, characterization and detergent compatibility of alkaline protease from soil isolate Bacillus cereus AG1. Int J Curr Microbiol App Sci 7(8):587–600. https://doi.org/10.20546/ijcmas.2018.708.064
Vijitra L, Yotchaisarn M, Saengha W et al (2019) Protease-producing bacteria from soil in Nasinuan community forest, Mahasarakham Province Thailand. Biomed Pharmacol 12(2):587–595. https://doi.org/10.13005/bpj/1678
Preeti S, Chayanika P, Anil K et al (2021) Microbial proteases: ubiquitous enzymes with innumerable uses. 3 Biotech 11:428. https://doi.org/10.1007/s13205-021-02928-z
Iuliia D, Margarita S (2020) The practical potential of Bacilli and their enzymes for industrial production. Front Microbiol 11:1782. https://doi.org/10.3389/fmicb.2020.01782
Fabiano J, de Melo Ricardo R, Sato Helia H (2017) An overview of Bacillus proteases: from production to application. Crit Rev Biotechnol 38(3):1549–780. https://doi.org/10.1080/07388551.2017.1354354
Zhen F, Yang-C JZ et al (2017) Keratinolytic protease: a green biocatalyst for leather industry. Appl Microbiol Biotechnol 101:7771–7779. https://doi.org/10.1007/s00253-017-8484-1
Yuhong H, Mateusz Ł, Alexander H et al (2020) Novel keratinolytic enzymes, discovered from a talented and efficient bacterial keratin degrader. Sci Rep 10:10033. https://doi.org/10.1038/s41598-020-66792-2
Renata B, Violeta V, Virgilijus V (2021) Enzymes for leather processing: effect on pickling and chroming. Materials 14(6):1480. https://doi.org/10.3390/2Fma14061480
Yeasmin A, Javed F, Aa H et al (2021) Keratinolytic protease from Pseudomonas aeruginosa for leather skin processing. JGEB 19:53. https://doi.org/10.1186/s43141-021-00149-8
Jayant P, Pallavi S, Il Patil et al (2015) Extraction of silver from waste x-ray films using protease enzyme. IJABR 6(2):220–226 (http://www.bipublication.com/)
Vaishali C (2013) Recovery of silver from used x-ray films by Aspergillus versicolor protease. JAIR 2(1):2278–5213
Amira H, Eida M (2016) Recovery of silver from used x-ray film using alkaline protease from Bacillus subtilis sub sp. Subtilis Afr J Biotechnol 15(26):1413–1416. https://doi.org/10.5897/AJB2016.15340
Ibrahim A, Elbadawi Y, Mohamed Al et al (2019) Alkaline serine protease from the new halotolerant alkaliphilic Salipaludibacillus agaradhaerens strain AK-R: purifcation and properties. Biotech 9(11):391. https://doi.org/10.1007/2Fs13205-019-1928-9
Tamilvendan M, Arulmani M, Rl S et al (2020) Identification of a novel thermostable alkaline protease from Bacillus megaterium-TK1 for the detergent and leather industry. Biology 9:472. https://doi.org/10.3390/biology9120472
Yuanliang H, Dan Y, Wang Z et al (2019) Purification and characterization of a novel, highly potent fbrinolytic enzyme from Bacillus subtilis DC27 screened from Douchi, a traditional Chinese fermented soybean food. Nature- Sci Rep 9:9235. https://doi.org/10.1038/s41598-019-45686-y
Farwa A, Shourong W, Kasim V (2021) Role of fibrinolytic enzymes in anti-thrombosis therapy. Front Mol Biosci 8:680397. https://doi.org/10.3389/fmolb.2021.680397
Fathma S, Elisabeth K, Pl N et al (2020) Fibrinolytic bacteria of Indonesian fermented soybean: preliminary study on enzyme activity and protein profile, Food Sci. Technol, Campinas 40(2):458–465. https://doi.org/10.1590/fst.23919
Rasika P, Vasudeo Z, Siddhivinayak B et al (2009) Application of protease isolated from Bacillus sp 158 in enzymatic cleansing of contact lenses. Biotechnol J 8(2):276–280. https://doi.org/10.3923/biotech.2009.276.280
Olga M, Ayslu M, Vladimir E et all (2017) Effects of Bacillus serine proteases on the bacterial biofilms. BioMed Res Int 8525912. https://doi.org/10.1155/2017/8525912
Janos A, Ferenc T, Jozsef T (2013) Research applications of proteolytic enzymes in molecular biology. Biomolecules 3:923–942. https://doi.org/10.3390/biom3040923
Nasir A, Rita D, Alexandre D et all (2017) Current Nucleic Acid Extraction Methods and Their Implications to Point-of-Care Diagnostics. Biomed Res Int 2017:9306564. https://doi.org/10.1155/2017/9306564
Swati J, Satyanarayana T (2013) Characteristics and applications of a recombinant alkaline serine protease from a novel bacterium Bacillus lehensis. Bioresour Technol 131:76–85. https://doi.org/10.1016/j.biortech.2012.12.124
Kim J, Kwon MiYeon, Kim S (2016) Biological degumming of silk fabrics with proteolytic enzymes. J Nat Fibers 13(6):629–639. https://doi.org/10.1080/15440478.2015.1093578
Lei Z, Junxiong LP et al (2022) Recent advances in environmentally friendly and green degumming processes of silk for textile and non-textile applications. Polymers 14:659. https://doi.org/10.3390/polym14040659
Sneha U, Poornima R, Sridhar S (2014) Optimization and decolorization of malachite green using Pseudomonas putida. J Chem Pharm Res 6(12):50–57
Anson ML (1938) The estimation of pepsin, trypsin, papain, and cathepsin with haemoglobin. J Gen Physiol 22(1):79–89. https://doi.org/10.1085/jgp.22.1.79
Chandran M, Getachew G, Mesfin T (2021) Isolation, screening, characterization, and identification of alkaline protease producing bacteria from leather industry effluent. Ann Microbiology 71:24. https://doi.org/10.1186/s13213-021-01631-x
Naveena BM, Mendiratta SK, Anjaneyulu ASR (2004) Tenderization of buffalo meat using plant proteases from Cucumis trigonus Roxb (Kachri) and Zingiber officinale roscoe (Ginger rhizome). Meat Sci 68(3):363–369. https://doi.org/10.1016/j.meatsci.2004.04.004
Furtak K, Gajda AM (2018) Activity and Variety of Soil Microorganisms Depending on the Diversity of the Soil Tillage System [Internet]. Sustainability of Agroecosystems. InTech 2018. Available from: https://doi.org/10.5772/intechopen.72966
Saxena A, Kumar M, Chakdar H et al (2019) Bacillus species in soil as a natural resource for plant health and nutrition. J Appl Microbiol 128(6):1583–1594. https://doi.org/10.1111/jam.14506
Prashant S, Shilpa A, Kiransinh N et al (2020) Thermostable alkaline proteases from bacteria: A review. NCIBS 978(93):5407–322-9. https://doi.org/10.2139/ssrn.3562387
Harwood CR, Yoshimi K (2022) The ins and outs of Bacillus proteases: activities, functions and commercial significance. FEMS Microbiol Rev 46(1):1–20. https://doi.org/10.1093/femsre/fuab046
Mayuri S, Yogesh G, Shalini A et al (2019) A review on microbial alkaline protease: an essential tool for various industrial approaches. Ind Biotechnol 15(2):69–78. https://doi.org/10.1089/ind.2018.0032
Rayan S, Elham M, Saeed A (2021) Cohnella 1759 cysteine protease shows significant long term half-life and impressive increased activity in presence of some chemical reagents. Sci Rep 11:4573. https://doi.org/10.1038/2Fs41598-021-84267-w
Juan L, Anupama S, Marie J et al (2018) Papain-like cysteine proteases in Carica papaya: lineage-specific gene duplication and expansion. BMC Genomics 19:26 (https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-017-4394-y#citeas)
Leila K, Mohammad Y, Mehdi I (2020) The effects of pH and temperature on cysteine protease (cathepsin B) activity in miracidia and eggs of Fasciola hepatica. Iran J Parasitol 15(2):233–239 (http://ijpa.tums.ac.ir)
Sarika M, Manisha A, Aishwarya P (2018) Isolation, partial purification, biochemical characterization and detergent compatibility of alkaline protease produced by Bacillus subtilis, Alcaligenes faecalis and Pseudomonas aeruginosa obtained from sea water samples. JGEB 16(1):39–46. https://doi.org/10.1016/j.jgeb.2017.10.001
Gurumallesh P, Alagu K et al (2019) A systematic reconsideration on proteases. Int J of Biol Macromol 128:254–267. https://doi.org/10.1016/j.ijbiomac.2019.01.081
Bajaj BK, Sharma P (2011) An alkali-thermotolerant extracellular protease from a newly isolated Streptomyces sp. DP2. New Biotech 28(6):725–732. https://doi.org/10.1016/j.nbt.2011.01.001
James David Morton, Zuhaib Fayaz Bhat, Alaa El-Din Ahmed Bekhit (2019) Proteases and meat tenderization. Encyof Food Chem 2:309–313. https://doi.org/10.1016/B978-0-08-100596-5.21663-6.
Funding
This work has been partially funded by OU-DST-PURSE intramural grant and RUSA 2.0.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Responsible Editor: Gisele Monteiro
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Keshapaga, U.R., Jathoth, K., Singh, S.S. et al. Characterization of high-yield Bacillus subtilis cysteine protease for diverse industrial applications. Braz J Microbiol 54, 739–752 (2023). https://doi.org/10.1007/s42770-023-00992-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-023-00992-6