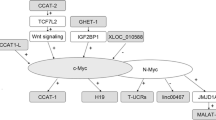
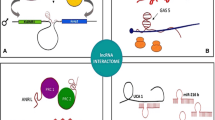
Abstract
Recent advancements in genomic and computational technologies have identified numerous untranslated noncoding transcripts including long noncoding RNAs (lncRNAs) in the human genome. Subsequently, the abundance and functional diversity of lncRNAs have been characterized in cellular and pathological pathways of various diseases and cancer. The transcription factor MYC is one of the most pivotal oncogenes, and the oncogenic pathways of MYC have been thought to be competently elucidated. However, recent studies implicating lncRNAs in the MYC pathways show that our research endeavors and knowledge to this day do not thoroughly elaborate the MYC pathways in cancer. It has been shown in recent studies that lncRNAs employ novel mechanisms to tune MYC activity and orchestrate MYC pathways. Furthermore, a number of lncRNAs that have not been dissected have been profiled and identified in the oncogenic MYC pathways. Thus, the research focusing on lncRNAs provides novel biological knowledge of the MYC pathways and will eventually lead to new potential biomarkers and therapeutic targets in MYC-driven cancers. This review discusses the various functional mechanisms of MYC-associated lncRNAs in MYC activity and MYC pathways.




Similar content being viewed by others
References
Amati, B., Alevizopoulos, K., & Vlach, J. (1998). Myc and the cell cycle. Frontiers in Bioscience, 3, d250–d268.
Ariel, I., de Groot, N., & Hochberg, A. (2000). Imprinted H19 gene expression in embryogenesis and human cancer: the oncofetal connection. American Journal of Medical Genetics, 91, 46–50.
Arima, T., Matsuda, T., Takagi, N., & Wake, N. (1997). Association of IGF2 and H19 imprinting with choriocarcinoma development. Cancer Genetics and Cytogenetics, 93, 39–47.
Arnold, P. R., Wells, A. D., & Li, X. C. (2019). Diversity and emerging roles of enhancer RNA in regulation of gene expression and cell fate. Front Cell Dev Biol, 7, 377.
Atmadibrata, B., Liu, P. Y., Sokolowski, N., Zhang, L., Wong, M., Tee, A. E., et al. (2014). The novel long noncoding RNA linc00467 promotes cell survival but is down-regulated by N-Myc. PLoS ONE, 9, e88112.
Banet, G., Bibi, O., Matouk, I., Ayesh, S., Laster, M., Kimber, K. M., et al. (2000). Characterization of human and mouse H19 regulatory sequences. Molecular Biology Reports, 27, 157–165.
Bernard, S., & Eilers, M. (2006). Control of cell proliferation and growth by Myc proteins. Results and Problems in Cell Differentiation, 42, 329–342.
Blackwood, E. M., & Eisenman, R. N. (1991). Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science, 251, 1211–1217.
Boissonnas, C. C., Abdalaoui, H. E., Haelewyn, V., Fauque, P., Dupont, J. M., Gut, I., et al. (2010). Specific epigenetic alterations of IGF2-H19 locus in spermatozoa from infertile men. European Journal of Human Genetics, 18, 73–80.
Brown, S. D. (1991). XIST and the mapping of the X chromosome inactivation centre. BioEssays, 13, 607–612.
Calin, G. A., & Croce, C. M. (2006). MicroRNA signatures in human cancers. Nature Reviews Cancer, 6, 857–866.
Calin, G. A., Dumitru, C. D., Shimizu, M., Bichi, R., Zupo, S., Noch, E., et al. (2002). Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A, 99, 15524–15529.
Cao, L., Zhang, P., Li, J., & Wu, M. (2017). LAST, a c-Myc-inducible long noncoding RNA, cooperates with CNBP to promote CCND1 mRNA stability in human cells. ELIFE, 6, e30433.
Castelnuovo, M., Massone, S., Tasso, R., Fiorino, G., Gatti, M., Robello, M., et al. (2010). An Alu-like RNA promotes cell differentiation and reduces malignancy of human neuroblastoma cells. The FASEB Journal, 24, 4033–4046.
Chang, T. C., Yu, D., Lee, Y. S., Wentzel, E. A., Arking, D. E., West, K. M., et al. (2008). Widespread microRNA repression by Myc contributes to tumorigenesis. Nature Genetics, 40, 43–50.
Chen, H., Liu, H., & Qing, G. (2018). Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduction and Targeted Therapy, 3, 5.
Chen, B., Dragomir, M. P., Fabris, L., Bayraktar, R., Knutsen, E., Liu, X., et al. (2020). The long noncoding RNA CCAT2 induces chromosomal instability through BOP1-AURKB signaling. Gastroenterology, 159, 2146–2162.
Cobbold, L. C., Spriggs, K. A., Haines, S. J., Dobbyn, H. C., Hayes, C., de Moor, C. H., et al. (2008). Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Molecular and Cellular Biology, 28, 40–49.
Cocchi, G., Marsico, C., Cosentino, A., Spadoni, C., Rocca, A., De Crescenzo, A., & Riccio, A. (2013). Silver-Russell syndrome due to paternal H19/IGF2 hypomethylation in a twin girl born after in vitro fertilization. American Journal of Medical Genetics Part A, 161A, 2652–2655.
Colombo, T., Farina, L., Macino, G., & Paci, P. (2015). PVT1: a rising star among oncogenic long noncoding RNAs. BioMed Research International, 2015, 304208.
Dalla-Favera, R., Bregni, M., Erikson, J., Patterson, D., Gallo, R. C., & Croce, C. M. (1982a). Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proceedings of the National Academy of Sciences of the United States of America, 79, 7824–7827.
Dalla-Favera, R., Gelmann, E. P., Martinotti, S., Franchini, G., Papas, T. S., Gallo, R. C., & Wong-Staal, F. (1982b). Cloning and characterization of different human sequences related to the onc gene (v-myc) of avian myelocytomatosis virus (MC29). Proceedings of the National Academy of Sciences of the United States of America, 79, 6497–6501.
Dang, C. V. (2012). MYC on the path to cancer. Cell, 149, 22–35.
DeBaun, M. R., Niemitz, E. L., McNeil, D. E., Brandenburg, S. A., Lee, M. P., & Feinberg, A. P. (2002). Epigenetic alterations of H19 and LIT1 distinguish patients with Beckwith-Wiedemann syndrome with cancer and birth defects. American Journal of Human Genetics, 70, 604–611.
Dinescu, S., Ignat, S., Lazar, A. D., Constantin, C., Neagu, M., & Costache, M. (2019). Epitranscriptomic signatures in lncRNAs and their possible roles in cancer. Genes (Basel), 10, 52.
Dong, Y. X., Pang, Z. G., Zhang, J. C., Hu, J. Q., & Wang, L. Y. (2019). Long non-coding RNA GClnc1 promotes progression of colorectal cancer by inhibiting p53 signaling pathway. European Review for Medical and Pharmacological Sciences, 23, 5705–5713.
Doose, G., Haake, A., Bernhart, S. H., Lopez, C., Duggimpudi, S., Wojciech, F., et al. (2015). MINCR is a MYC-induced lncRNA able to modulate MYC’s transcriptional network in Burkitt lymphoma cells. Proceedings of the National Academy of Sciences of the United States of America, 112, E5261–E5270.
Doose, G., Hoffmann, S., & Iaccarino, I. (2016). Reply to Hart et al.: MINCR and MYC: More than expression correlation. Proceedings of the National Academy of Sciences of the United States of America, 113, E498.
Doyle, L. A., Yang, W., Rishi, A. K., Gao, Y., & Ross, D. D. (1996). H19 gene overexpression in atypical multidrug-resistant cells associated with expression of a 95-kilodalton membrane glycoprotein. Cancer Research, 56, 2904–2907.
Duesberg, P. H., Bister, K., & Vogt, P. K. (1977). The RNA of avian acute leukemia virus MC29. Proceedings of the National Academy of Sciences of the United States of America, 74, 4320–4324.
Eilers, M. (1999). Control of cell proliferation by Myc family genes. Molecules and Cells, 9, 1–6.
El-Naggar, A. K., Lai, S., Tucker, S. A., Clayman, G. L., Goepfert, H., Hong, W. K., & Huff, V. (1999). Frequent loss of imprinting at the IGF2 and H19 genes in head and neck squamous carcinoma. Oncogene, 18, 7063–7069.
Feng, J., Ma, J., Liu, S., Wang, J., & Chen, Y. (2019). A noncoding RNA LINC00504 interacts with c-Myc to regulate tumor metabolism in colon cancer. Journal of Cellular Biochemistry, 120, 14725–14734.
Feng, Y. C., Liu, X. Y., Teng, L., Ji, Q., Wu, Y., Li, J. M., et al. (2020). c-Myc inactivation of p53 through the pan-cancer lncRNA MILIP drives cancer pathogenesis. Nature Communications, 11, 4980.
Frith, M. C., Forrest, A. R., Nourbakhsh, E., Pang, K. C., Kai, C., Kawai, J., et al. (2006). The abundance of short proteins in the mammalian proteome. PLoS Genetics, 2, e52.
Gao, Z. H., Suppola, S., Liu, J., Heikkila, P., Janne, J., & Voutilainen, R. (2002). Association of H19 promoter methylation with the expression of H19 and IGF-II genes in adrenocortical tumors. Journal of Clinical Endocrinology and Metabolism, 87, 1170–1176.
Garcia-Gutierrez, L., Delgado, M. D., & Leon, J. (2019). MYC oncogene contributions to release of cell cycle brakes. Genes (Basel), 10, 244.
Guo, X., & Hua, Y. (2017). CCAT1: An oncogenic long noncoding RNA in human cancers. Journal of Cancer Research and Clinical Oncology, 143, 555–562.
Hart, J. R., Roberts, T. C., Weinberg, M. S., Morris, K. V., & Vogt, P. K. (2014). MYC regulates the non-coding transcriptome. Oncotarget, 5, 12543–12554.
Hart, J. R., Weinberg, M. S., Morris, K. V., Roberts, T. C., Janda, K. D., Garner, A. L., & Vogt, P. K. (2016). MINCR is not a MYC-induced lncRNA. Proceedings of the National Academy of Sciences of the United States of America, 113, E496–E497.
He, X., Tan, X., Wang, X., Jin, H., Liu, L., Ma, L., et al. (2014). C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumour Biology, 35, 12181–12188.
He, J., Li, F., Zhou, Y., Hou, X., Liu, S., Li, X., et al. (2020a). LncRNA XLOC_006390 promotes pancreatic carcinogenesis and glutamate metabolism by stabilizing c-Myc. Cancer Letters, 469, 419–428.
He, Y., Shi, Q., Zhang, Y., Yuan, X., & Yu, Z. (2020b). Transcriptome-wide 5-methylcytosine functional profiling of long non-coding RNA in hepatocellular carcinoma. Cancer Management and Research, 12, 6877–6885.
Horos, R., Buscher, M., Kleinendorst, R., Alleaume, A. M., Tarafder, A. K., Schwarzl, T., et al. (2019). The small non-coding vault RNA1-1 acts as a riboregulator of autophagy. Cell, 176, 1054–1067.
Hsieh, A. L., Walton, Z. E., Altman, B. J., Stine, Z. E., & Dang, C. V. (2015). MYC and metabolism on the path to cancer. Seminars in Cell & Developmental Biology, 43, 11–21.
Hu, T., & Lu, Y. R. (2015). BCYRN1, a c-MYC-activated long non-coding RNA, regulates cell metastasis of non-small-cell lung cancer. Cancer Cell International, 15, 36.
Hu, G., Lou, Z., & Gupta, M. (2014). The long non-coding RNA GAS5 cooperates with the eukaryotic translation initiation factor 4E to regulate c-Myc translation. PLoS ONE, 9, e107016.
Hu, Y., Wang, F., Xu, F., Fang, K., Fang, Z., Shuai, X., et al. (2020). A reciprocal feedback of Myc and lncRNA MTSS1-AS contributes to extracellular acidity-promoted metastasis of pancreatic cancer. Theranostics, 10, 10120–10140.
Huang, J. Z., Chen, M., Chen, G. X. C., Zhu, S., Huang, H., Hu, M., et al. (2017). A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Molecular Cell, 68, 171–184.
Huarte, M. (2015). The emerging role of lncRNAs in cancer. Nature Medicine, 21, 1253–1261.
Hung, C. L., Wang, L. Y., Yu, Y. L., Chen, H. W., Srivastava, S., Petrovics, G., & Kung, H. J. (2014). A long noncoding RNA connects c-Myc to tumor metabolism. Proceedings of the National Academy of Sciences of the United States of America, 111, 18697–18702.
Hurst, L. D., & Smith, N. G. (1999). Molecular evolutionary evidence that H19 mRNA is functional. Trends in Genetics, 15, 134–135.
Ingolia, N. T., Lareau, L. F., & Weissman, J. S. (2011). Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell, 147, 789–802.
Ji, Z., Song, R., Regev, A., & Struhl, K. (2015). Many lncRNAs, 5’UTRs, and pseudogenes are translated and some are likely to express functional proteins. ELIFE, 4, e8890.
Kawasaki, Y., Komiya, M., Matsumura, K., Negishi, L., Suda, S., Okuno, M., et al. (2016). MYU, a target lncRNA for Wnt/c-Myc signaling, mediates induction of CDK6 to promote cell cycle progression. Cell Report, 16, 2554–2564.
Kim, T., & Croce, C. M. (2018). Long noncoding RNAs: Undeciphered cellular codes encrypting keys of colorectal cancer pathogenesis. Cancer Letters, 417, 89–95.
Kim, T., Cui, R., Jeon, Y. J., Lee, J. H., Lee, J. H., Sim, H., et al. (2014). Long-range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARLo-5. Proceedings of the National Academy of Sciences of the United States of America, 111, 4173–4178.
Kim, T., Jeon, Y. J., Cui, R., Lee, J. H., Peng, Y., Kim, S. H., et al. (2015a). Role of MYC-regulated long noncoding RNAs in cell cycle regulation and tumorigenesis. Journal of the National Cancer Institute, 107, 505.
Kim, T., Cui, R., Jeon, Y. J., Fadda, P., Alder, H., & Croce, C. M. (2015b). MYC-repressed long noncoding RNAs antagonize MYC-induced cell proliferation and cell cycle progression. Oncotarget, 6, 18780–18789.
Kohl, N. E., Kanda, N., Schreck, R. R., Bruns, G., Latt, S. A., Gilbert, F., & Alt, F. W. (1983). Transposition and amplification of oncogene-related sequences in human neuroblastomas. Cell, 35, 359–367.
Li, J., Xuan, Z., & Liu, C. (2013). Long non-coding RNAs and complex human diseases. International Journal of Molecular Sciences, 14, 18790–18808.
Li, Y., Wang, Z., Shi, H., Li, H., Li, L., Fang, R., et al. (2016). HBXIP and LSD1 scaffolded by lncRNA Hotair mediate transcriptional activation by c-Myc. Cancer Research, 76, 293–304.
Li, S., Zhang, S., & Chen, J. (2019). c-Myc induced upregulation of long non-coding RNA SNHG16 enhances progression and carcinogenesis in oral squamous cell carcinoma. Cancer Gene Therapy, 26, 400–410.
Li, H., Zeng, Z., Yang, X., Chen, Y., He, L., & Wan, T. (2020). LncRNA GClnc1 may contribute to the progression of ovarian cancer by regulating p53 signaling pathway. European Journal of Histochemistry, 64, 3166.
Li, J., Liao, T., Liu, H., Yuan, H., Ouyang, T., Wang, J., et al. (2021). Hypoxic glioma stem cell-derived exosomes containing Linc01060 promote progression of glioma by regulating the MZF1/c-Myc/HIF-1alpha. Cancer Research, 81, 114–128.
Liang, J., Zhou, H., Gerdt, C., Tan, M., Colson, T., Kaye, K. M., et al. (2016). Epstein-Barr virus super-enhancer eRNAs are essential for MYC oncogene expression and lymphoblast proliferation. Proceedings of the National Academy of Sciences of the United States of America, 113, 14121–14126.
Liao, L. M., Sun, X. Y., Liu, A. W., Wu, J. B., Cheng, X. L., Lin, J. X., et al. (2014). Low expression of long noncoding XLOC_010588 indicates a poor prognosis and promotes proliferation through upregulation of c-Myc in cervical cancer. Gynecologic Oncology, 133, 616–623.
Liao, Y., Jung, S. H., & Kim, T. (2020). A-to-I RNA editing as a tuner of noncoding RNAs in cancer. Cancer Letters, 494, 88–93.
Lin, C. Y., Loven, J., Rahl, P. B., Paranal, R. M., Burge, C. B., Bradner, J. E., et al. (2012). Transcriptional amplification in tumor cells with elevated c-Myc. Cell, 151, 56–67.
Ling, H., Spizzo, R., Atlasi, Y., Nicoloso, M., Shimizu, M., Redis, R. S., et al. (2013). CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Research, 23, 1446–1461.
Liu, H., Xu, Y., Yao, B., Sui, T., Lai, L., & Li, Z. (2020a). A novel N6-methyladenosine (m6A)-dependent fate decision for the lncRNA THOR. Cell Death and Disease, 11, 613.
Liu, J., Xu, R., Mai, S. J., Ma, Y. S., Zhang, M. Y., Cao, P. S., et al. (2020b). LncRNA CSMD1-1 promotes the progression of Hepatocellular Carcinoma by activating MYC signaling. Theranostics, 10, 7527–7544.
Liu, S., Zheng, Y., Zhang, Y., Zhang, J., Xie, F., Guo, S., et al. (2020c). Methylation-mediated LINC00261 suppresses pancreatic cancer progression by epigenetically inhibiting c-Myc transcription. Theranostics, 10, 10634–10651.
Lu, Y., Hu, Z., Mangala, L. S., Stine, Z. E., Hu, X., Jiang, D., et al. (2018). MYC targeted long noncoding RNA DANCR promotes cancer in part by reducing p21 levels. Cancer Research, 78, 64–74.
Luan, S., Luo, J., Liu, H., & Li, Z. (2019). Regulation of RNA decay and cellular function by 3’-5’ exoribonuclease DIS3L2. RNA Biology, 16, 160–165.
Luo, L., Tang, H., Ling, L., Li, N., Jia, X., Zhang, Z., et al. (2018). LINC01638 lncRNA activates MTDH-Twist1 signaling by preventing SPOP-mediated c-Myc degradation in triple-negative breast cancer. Oncogene, 37, 6166–6179.
Ma, F., Liu, X., Zhou, S., Li, W., Liu, C., Chadwick, M., & Qian, C. (2019). Long non-coding RNA FGF13-AS1 inhibits glycolysis and stemness properties of breast cancer cells through FGF13-AS1/IGF2BPs/Myc feedback loop. Cancer Letters, 450, 63–75.
Matsumoto, A., Pasut, A., Matsumoto, M., Yamashita, R., Fung, J., Monteleone, E., et al. (2017). mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature, 541, 228–232.
Menezes, M. R., Balzeau, J., & Hagan, J. P. (2018). 3’ RNA uridylation in epitranscriptomics, gene regulation, and disease. Frontiers in Molecular Biosciences, 5, 61.
Nau, M. M., Brooks, B. J., Battey, J., Sausville, E., Gazdar, A. F., Kirsch, I. R., et al. (1985). L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature, 318, 69–73.
Nie, Z., Hu, G., Wei, G., Cui, K., Yamane, A., Resch, W., et al. (2012). c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell, 151, 68–79.
Nissan, A., Stojadinovic, A., Mitrani-Rosenbaum, S., Halle, D., Grinbaum, R., Roistacher, M., et al. (2012). Colon cancer associated transcript-1: a novel RNA expressed in malignant and pre-malignant human tissues. International Journal of Cancer, 130, 1598–1606.
O’Donnell, K. A., Wentzel, E. A., Zeller, K. I., Dang, C. V., & Mendell, J. T. (2005). c-Myc-regulated microRNAs modulate E2F1 expression. Nature, 435, 839–843.
Olivero, C. E., Martinez-Terroba, E., Zimmer, J., Liao, C., Tesfaye, E., Hooshdaran, N., et al. (2020). p53 Activates the long noncoding RNA Pvt1b to inhibit Myc and suppress tumorigenesis. Molecular Cell, 77, 761–774.
Ouyang, D., Li, R., Li, Y., & Zhu, X. (2019). Construction of a competitive endogenous RNA network in uterine corpus endometrial carcinoma. Medical Science Monitor, 25, 7998–8010.
Pisani, G., & Baron, B. (2020). NEAT1 and paraspeckles in cancer development and chemoresistance. Noncoding RNA, 6, 43.
Raffeiner, P., Hart, J. R., Garcia-Caballero, D., Bar-Peled, L., Weinberg, M. S., & Vogt, P. K. (2020). An MXD1-derived repressor peptide identifies noncoding mediators of MYC-driven cell proliferation. Proceedings of the National Academy of Sciences of the United States of America, 117, 6571–6579.
Raveh, E., Matouk, I. J., Gilon, M., & Hochberg, A. (2015). The H19 Long non-coding RNA in cancer initiation, progression and metastasis—a proposed unifying theory. Molecular Cancer, 14, 184.
Rinn, J. L., Kertesz, M., Wang, J. K., Squazzo, S. L., Xu, X., Brugmann, S. A., et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell, 129, 1311–1323.
Ruiz-Orera, J., Messeguer, X., Subirana, J. A., & Alba, M. M. (2014). Long non-coding RNAs as a source of new peptides. ELIFE, 3, e3523.
Sahu, D., Ho, S. Y., Juan, H. F., & Huang, H. C. (2018). High-risk, expression-based prognostic long noncoding rna signature in neuroblastoma. JNCI Cancer Spectrum, 2, y15.
Salmena, L., Poliseno, L., Tay, Y., Kats, L., & Pandolfi, P. P. (2011). A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell, 146, 353–358.
Sampson, V. B., Rong, N. H., Han, J., Yang, Q., Aris, V., Soteropoulos, P., et al. (2007). MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Research, 67, 9762–9770.
Shahabi, S., Kumaran, V., Castillo, J., Cong, Z., Nandagopal, G., Mullen, D. J., et al. (2019). LINC00261 is an epigenetically regulated tumor suppressor essential for activation of the DNA damage response. Cancer Research, 79, 3050–3062.
Shen, W., Liang, X. H., Sun, H., De Hoyos, C. L., & Crooke, S. T. (2017). Depletion of NEAT1 lncRNA attenuates nucleolar stress by releasing sequestered P54nrb and PSF to facilitate c-Myc translation. PLoS ONE, 12, e173494.
Shi, X., Cui, Z., Liu, X., Wu, S., Wu, Y., Fang, F., & Zhao, H. (2019). LncRNA FIRRE is activated by MYC and promotes the development of diffuse large B-cell lymphoma via Wnt/beta-catenin signaling pathway. Biochemical and Biophysical Research Communications, 510, 594–600.
Shigeyasu, K., Toden, S., Ozawa, T., Matsuyama, T., Nagasaka, T., Ishikawa, T., et al. (2020). The PVT1 lncRNA is a novel epigenetic enhancer of MYC, and a promising risk-stratification biomarker in colorectal cancer. Molecular Cancer, 19, 155.
Shima, H., & Igarashi, K. (2020). N 1-methyladenosine (m1A) RNA modification: The key to ribosome control. Journal of Biochemistry, 167, 535–539.
Sui, J., Miao, Y., Han, J., Nan, H., Shen, B., Zhang, X., et al. (2018a). Systematic analyses of a novel lncRNA-associated signature as the prognostic biomarker for Hepatocellular Carcinoma. Cancer Medicine, 7, 3240.
Sui, Y., Han, Y., Zhao, X., Li, D., & Li, G. (2018b). Long non-coding RNA GClnc1 promotes tumorigenesis in osteosarcoma by inhibiting p53 signaling. Biochemical and Biophysical Research Communications, 507, 36–42.
Sun, T. T., He, J., Liang, Q., Ren, L. L., Yan, T. T., Yu, T. C., et al. (2016). LncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discovery, 6, 784–801.
Sun, Y., Jin, S. D., Zhu, Q., Han, L., Feng, J., Lu, X. Y., et al. (2017). Long non-coding RNA LUCAT1 is associated with poor prognosis in human non-small lung cancer and regulates cell proliferation via epigenetically repressing p21 and p57 expression. Oncotarget, 8, 28297–28311.
Sun, Q., Hao, Q., & Prasanth, K. V. (2018). Nuclear long noncoding RNAs: Key regulators of gene expression. Trends in Genetics, 34, 142–157.
Takeuchi, S., Hofmann, W. K., Tsukasaki, K., Takeuchi, N., Ikezoe, T., Matsushita, M., et al. (2007). Loss of H19 imprinting in adult T-cell leukaemia/lymphoma. British Journal of Haematology, 137, 380–381.
Tang, J., Yan, T., Bao, Y., Shen, C., Yu, C., Zhu, X., et al. (2019). LncRNA GLCC1 promotes colorectal carcinogenesis and glucose metabolism by stabilizing c-Myc. Nature Communications, 10, 3499.
Tanos, V., Prus, D., Ayesh, S., Weinstein, D., Tykocinski, M. L., De-Groot, N., et al. (1999). Expression of the imprinted H19 oncofetal RNA in epithelial ovarian cancer. European Journal of Obstetrics, Gynecology, and Reproductive Biology, 85, 7–11.
Tauber, H., Huttelmaier, S., & Kohn, M. (2019). POLIII-derived non-coding RNAs acting as scaffolds and decoys. J Molecular Cell Biology, 11, 880–885.
Tran, D., Kessler, C., Niehus, S. E., Mahnkopf, M., Koch, A., & Tamura, T. (2018). Myc target gene, long intergenic noncoding RNA, Linc00176 in hepatocellular carcinoma regulates cell cycle and cell survival by titrating tumor suppressor microRNAs. Oncogene, 37, 75–85.
Tseng, Y. Y., Moriarity, B. S., Gong, W., Akiyama, R., Tiwari, A., Kawakami, H., et al. (2014). PVT1 dependence in cancer with MYC copy-number increase. Nature, 512, 82–86.
Viswanathan, S. R., Daley, G. Q., & Gregory, R. I. (2008). Selective blockade of microRNA processing by Lin28. Science, 320, 97–100.
Vogt, P. K. (2012). Retroviral oncogenes: A historical primer. Nature Reviews Cancer, 12, 639–648.
Wang, K. C., & Chang, H. Y. (2011). Molecular mechanisms of long noncoding RNAs. Molecular Cell, 43, 904–914.
Wang, K. C., Yang, Y. W., Liu, B., Sanyal, A., Corces-Zimmerman, R., Chen, Y., et al. (2011). A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature, 472, 120–124.
Wang, O., Yang, F., Liu, Y., Lv, L., Ma, R., Chen, C., et al. (2017). C-MYC-induced upregulation of lncRNA SNHG12 regulates cell proliferation, apoptosis and migration in triple-negative breast cancer. American Journal of Translational Research, 9, 533–545.
Wang, Z., Yang, B., Zhang, M., Guo, W., Wu, Z., Wang, Y., et al. (2018). lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell, 33, 706–720.
Wang, C., Yang, Y., Zhang, G., Li, J., Wu, X., Ma, X., et al. (2019a). Long noncoding RNA EMS connects c-Myc to cell cycle control and tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America, 116, 14620–14629.
Wang, L., Cho, K. B., Li, Y., Tao, G., Xie, Z., & Guo, B. (2019b). Long noncoding RNA (lncRNA)-mediated competing endogenous rna networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. International Journal of Molecular Sciences, 20, 5758.
Wang, Z., Zhang, J., Yang, B., Li, R., Jin, L., Wang, Z., et al. (2019c). Long intergenic noncoding RNA 00261 acts as a tumor suppressor in non-small cell lung cancer via regulating miR-105/FHL1 axis. Journal of Cancer, 10, 6414–6421.
Wilusz, J. E., Freier, S. M., & Spector, D. L. (2008). 3’ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell, 135, 919–932.
Wrana, J. L. (1994). H19, a tumour suppressing RNA? BioEssays, 16, 89–90.
Wu, M., & Shen, J. (2019). From super-enhancer non-coding RNA to Immune checkpoint: frameworks to functions. Frontiers in Oncology, 9, 1307.
Wu, P., Mo, Y., Peng, M., Tang, T., Zhong, Y., Deng, X., et al. (2020a). Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Molecular Cancer, 19, 22.
Wu, R., Li, L., Bai, Y., Yu, B., Xie, C., Wu, H., et al. (2020b). The long noncoding RNA LUCAT1 promotes colorectal cancer cell proliferation by antagonizing Nucleolin to regulate MYC expression. Cell Death Dis, 11, 908.
Xin, Y., Li, Z., Shen, J., Chan, M. T., & Wu, W. K. (2016). CCAT1: A pivotal oncogenic long non-coding RNA in human cancers. Cell Proliferation, 49, 255–260.
Xu, T. P., Ma, P., Wang, W. Y., Shuai, Y., Wang, Y. F., Yu, T., et al. (2019). KLF5 and MYC modulated LINC00346 contributes to gastric cancer progression through acting as a competing endogeous RNA and indicates poor outcome. Cell Death and Differentiation, 26, 2179–2193.
Yang, F., Xue, X., Zheng, L., Bi, J., Zhou, Y., Zhi, K., et al. (2014). Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS Journal, 281, 802–813.
Yang, Z., Zhang, T., Han, S., Kusumanchi, P., Huda, N., Jiang, Y., & Liangpunsakul, S. (2020). Long noncoding RNA H19—a new player in the pathogenesis of liver diseases. Translational Research-Journal, S1931–5244, 30281–30284.
Yeganeh, M., & Hernandez, N. (2020). RNA polymerase III transcription as a disease factor. Genes & Development, 34, 865–882.
Yin, Q. F., Yang, L., Zhang, Y., Xiang, J. F., Wu, Y. W., Carmichael, G. G., & Chen, L. L. (2012). Long noncoding RNAs with snoRNA ends. Molecular Cell, 48, 219–230.
Yu, Y., Li, L., Zheng, Z., Chen, S., Chen, E., & Hu, Y. (2017). Long non-coding RNA linc00261 suppresses gastric cancer progression via promoting Slug degradation. Journal of Cellular and Molecular Medicine, 21, 955–967.
Zeng, C., Liu, S., Lu, S., Yu, X., Lai, J., Wu, Y., et al. (2018). The c-Myc-regulated lncRNA NEAT1 and paraspeckles modulate imatinib-induced apoptosis in CML cells. Molecular Cancer, 17, 130.
Zhang, E., Li, W., Yin, D., De, W., Zhu, L., Sun, S., & Han, L. (2016a). c-Myc-regulated long non-coding RNA H19 indicates a poor prognosis and affects cell proliferation in non-small-cell lung cancer. Tumour Biology, 37, 4007–4015.
Zhang, P., Cao, L., Fan, P., Mei, Y., & Wu, M. (2016b). LncRNA-MIF, a c-Myc-activated long non-coding RNA, suppresses glycolysis by promoting Fbxw7-mediated c-Myc degradation. EMBO Reports, 17, 1204–1220.
Zhang, X. Z., Liu, H., & Chen, S. R. (2020). Mechanisms of long non-coding RNAs in cancers and their dynamic regulations. Cancers (Basel), 12, 1245.
Zhuang, C., Ma, Q., Zhuang, C., Ye, J., Zhang, F., & Gui, Y. (2019). LncRNA GClnc1 promotes proliferation and invasion of bladder cancer through activation of MYC. The FASEB Journal, 33, 11045–11059.
Acknowledgements
The author thanks Dr. Peter K. Vogt for the review and scientific comments and Anja Zembrzycki for manuscript proofreading. This work was supported by the National Natural Science Foundation of China (NSFC) (Project No. 82073120).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The author declares no conflict of interest.
Rights and permissions
About this article
Cite this article
Kim, T. LncRNAs as key players in the MYC pathways. GENOME INSTAB. DIS. 2, 24–38 (2021). https://doi.org/10.1007/s42764-021-00032-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42764-021-00032-3