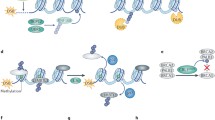
Abstract
The ubiquitin system plays a central role in diverse cellular processes including DNA damage response. As such, it is not surprising that its dysfunction contributes to various diseases including cancer and neurodegenerative disorders. An understanding of the ubiquitin system is, therefore, important in devising treatments for such diseases. In this review, we discuss the central role of ubiquitin in DNA damage response, specifically DNA double-strand break repair. We focus on recent findings on the role of ubiquitin in the DNA double-strand break repair pathway, possible nodes of modulation, and finally their implications for treatment of various diseases.




Similar content being viewed by others
References
Acs, K., et al. (2011). The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nature Structural & Molecular Biology,18, 1345.
Andreassen, P. R., D’Andrea, A. D., & Taniguchi, T. (2004). ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes & Development,18, 1958–1963.
Barton, O., et al. (2014). Polo-like kinase 3 regulates CtIP during DNA double-strand break repair in G1. Journal of Cell Biology,206, 877–894.
Bohgaki, M., et al. (2013). RNF168 ubiquitylates 53BP1 and controls its response to DNA double-strand breaks. Proceedings of the National Academy of Sciences,110, 20982–20987.
Brown, E. J., & Baltimore, D. (2000). ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes & Development,14, 397–402.
Brown, J. S., et al. (2015). Neddylation promotes ubiquitylation and release of Ku from DNA-damage sites. Cell Reports,11, 704–714.
Buisson, R., et al. (2017). Coupling of homologous recombination and the checkpoint by ATR. Molecular Cell,65, 336–346.
Bunting, S. F., et al. (2010). 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell,141, 243–254.
Butler, L. R., et al. (2012). The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response. The EMBO Journal,31, 3918–3934.
Callen, E., et al. (2013). 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell,153, 1266–1280.
Cao, J., & Yan, Q. (2012). Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Frontiers in Oncology,2, 26.
Centore, R. C., Yazinski, S. A., Tse, A., & Zou, L. (2012). Spartan/C1orf124, a reader of PCNA ubiquitylation and a regulator of UV-induced DNA damage response. Molecular Cell,46, 625–635.
Chau, V., et al. (1989). A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science,243, 1576–1583.
Chen, J., Feng, W., Jiang, J., Deng, Y., & Huen, M. S. (2012). Ring finger protein RNF169 antagonizes the ubiquitin-dependent signaling cascade at sites of DNA damage. Journal of Biological Chemistry,287, 27715–27722.
Chen, Z., et al. (2018). Ubiquitin-like protein FAT10 regulates DNA damage repair via modification of proliferating cell nuclear antigen. Molecular Medicine Reports,17, 7487–7496.
Chroma, K., et al. (2017). Tumors overexpressing RNF168 show altered DNA repair and responses to genotoxic treatments, genomic instability and resistance to proteotoxic stress. Oncogene,36, 2405–2422.
Clouaire, T., et al. (2018). Comprehensive mapping of histone modifications at DNA double-strand breaks deciphers repair pathway chromatin signatures. Molecular Cell,72, 250–262.e256.
Coleman, K. A., & Greenberg, R. A. (2011a). The BRCA1–RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. Journal of Biological Chemistry,286, 13669–13680.
Coleman, K. A., & Greenberg, R. A. (2011b). The BRCA1–RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. The Journal of biological chemistry,286, 13669–13680.
Connelly, J. C., de Leau, E. S., & Leach, D. R. (2003). Nucleolytic processing of a protein-bound DNA end by the E. coli SbcCD (MR) complex. DNA Repair,2, 795–807.
Cook, P. J., et al. (2009). Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature,458, 591.
DeJesus, R., et al. (2016). Functional CRISPR screening identifies the ufmylation pathway as a regulator of SQSTM1/p62. Elife,5, e17290.
Densham, R. M., et al. (2016). Human BRCA1–BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nature Structural & Molecular Biology,23, 647.
Deshaies, R. J., & Joazeiro, C. A. (2009). RING domain E3 ubiquitin ligases. Annual Review of Biochemistry,78, 399–434.
Deshpande, R. A., Lee, J.-H., Arora, S., & Paull, T. T. (2016). Nbs1 converts the human Mre11/Rad50 nuclease complex into an endo/exonuclease machine specific for protein–DNA adducts. Molecular Cell,64, 593–606.
Di Virgilio, M., et al. (2013). Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science,339, 711–715.
Doil, C., et al. (2009). RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell,136, 435–446.
Drané, P., et al. (2017). TIRR regulates 53BP1 by masking its histone methyl-lysine binding function. Nature,543, 211.
Elia, A. E. H., et al. (2015). Quantitative proteomic atlas of ubiquitination and acetylation in the DNA damage response. Molecular Cell,59, 867–881.
Falck, J., Coates, J., & Jackson, S. P. (2005). Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature,434, 605.
Feng, L., & Chen, J. (2012). The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nature Structural & Molecular Biology,19, 201.
Feng, L., et al. (2015). Cell cycle-dependent inhibition of 53BP1 signaling by BRCA1. Cell Discovery,1, 15019.
Ferretti, L. P., et al. (2016). Cullin3-KLHL15 ubiquitin ligase mediates CtIP protein turnover to fine-tune DNA-end resection. Nature Communications,7, 12628.
Fradet-Turcotte, A., et al. (2013). 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature,499, 50.
Galanty, Y., Belotserkovskaya, R., Coates, J., & Jackson, S. P. (2012). RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes & Development,26, 1179–1195.
Gatti, M., et al. (2015). RNF168 promotes noncanonical K27 ubiquitination to signal DNA damage. Cell Reports,10, 226–238.
Gudjonsson, T., et al. (2012). TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell,150, 697–709.
Guo, X., et al. (2017). Acetylation of 53BP1 dictates the DNA double strand break repair pathway. Nucleic Acids Research,46, 689–703.
Habisov, S., et al. (2016). Structural and functional analysis of a novel interaction motif within UFM1-activating enzyme 5 (UBA5) required for binding to ubiquitin-like proteins and ufmylation. Journal of Biological Chemistry,291, 9025–9041.
Han, X., et al. (2014). UbcH7 regulates 53BP1 stability and DSB repair. Proceedings of the National Academy of Sciences,111, 17456–17461.
Hatakeyama, S., Yada, M., Matsumoto, M., Ishida, N., & Nakayama, K.-I. (2001). U box proteins as a new family of ubiquitin-protein ligases. Journal of Biological Chemistry,276, 33111–33120.
Higashitsuji, H., et al. (2005). The oncoprotein gankyrin binds to MDM2/HDM2, enhancing ubiquitylation and degradation of p53. Cancer Cell,8, 75–87.
Hoa, N. N., et al. (2016). Mre11 is essential for the removal of lethal topoisomerase 2 covalent cleavage complexes. Molecular Cell,64, 580–592.
Hoege, C., Pfander, B., Moldovan, G.-L., Pyrowolakis, G., & Jentsch, S. (2002). RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature,419, 135.
Huang, Y.-F., Wee, S., Gunaratne, J., Lane, D. P., & Bulavin, D. V. (2014). Isg15 controls p53 stability and functions. Cell Cycle,13, 2199–2209.
Huang, L., et al. (1999). Structure of an E6AP–UbcH7 complex: Insights into ubiquitination by the E2–E3 enzyme cascade. Science,286, 1321–1326.
Huen, M. S., et al. (2007). RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell,131, 901–914.
Hurley, J. H., Lee, S., & Prag, G. (2006). Ubiquitin-binding domains. Biochemical Journal,399, 361–372.
Ilić, D., et al. (1998). Extracellular matrix survival signals transduced by focal adhesion kinase suppress p53-mediated apoptosis. The Journal of Cell Biology,143, 547–560.
Ismail, I. H., et al. (2015). The RNF138 E3 ligase displaces Ku to promote DNA end resection and regulate DNA repair pathway choice. Nature Cell Biology,17, 1446.
Jacquet, K., et al. (2016). The TIP60 complex regulates bivalent chromatin recognition by 53BP1 through direct H4K20me binding and H2AK15 acetylation. Molecular Cell,62, 409–421.
Jin, L., Williamson, A., Banerjee, S., Philipp, I., & Rape, M. (2008). Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell,133, 653–665.
Kato, K., et al. (2014). Fine-tuning of DNA damage-dependent ubiquitination by OTUB2 supports the DNA repair pathway choice. Molecular Cell,53, 617–630.
Kim, H., Chen, J., & Yu, X. (2007). Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science,316, 1202–1205.
Kim, H.-K., et al. (2014). The N-terminal methionine of cellular proteins as a degradation signal. Cell,156, 158–169.
Knies, K., et al. (2017). Biallelic mutations in the ubiquitin ligase RFWD3 cause Fanconi anemia. The Journal of Clinical Investigation,127, 3013–3027.
Kolas, N. K., et al. (2007). Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science,318, 1637–1640.
Komander, D., Clague, M. J., & Urbé, S. (2009). Breaking the chains: Structure and function of the deubiquitinases. Nature Reviews Molecular Cell Biology,10, 550.
Koyano, F., et al. (2014). Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature,510, 162.
Kumar, A., et al. (2014). ATR mediates a checkpoint at the nuclear envelope in response to mechanical stress. Cell,158, 633–646.
Lechtenberg, B. C., et al. (2016). Structure of a HOIP/E2~ ubiquitin complex reveals RBR E3 ligase mechanism and regulation. Nature,529, 546.
Lee, J.-H., & Paull, T. T. (2004). Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science,304, 93–96.
Lee, J.-H., & Paull, T. T. (2005). ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science,308, 551–554.
Lee, D.-H., et al. (2014). Dephosphorylation enables the recruitment of 53BP1 to double-strand DNA breaks. Molecular Cell,54, 512–525.
Lee, N. S., et al. (2018). Ring finger protein 126 (RNF126) suppresses ionizing radiation-induced p53-binding protein 1 (53BP1) focus formation. The Journal of Biological Chemistry,293, 588–598.
Li, T., Guan, J., Huang, Z., Hu, X., & Zheng, X. (2014). RNF168-mediated H2A neddylation antagonizes ubiquitylation of H2A and regulates DNA damage repair. Journal of Cell Science,127, 2238–2248.
Li, Y., et al. (2017). USP13 regulates the RAP80–BRCA1 complex dependent DNA damage response. Nature Communications,8, 15752.
Liu, C., et al. (2013). RNF168 forms a functional complex with RAD6 during the DNA damage response. Journal of Cell Science,126, 2042–2051.
Liu, H., et al. (2015). The deubiquitylating enzyme USP4 cooperates with CtIP in DNA double-strand break end resection. Cell Reports,13, 93–107.
Lou, Z., Minter-Dykhouse, K., Wu, X., & Chen, J. (2003). MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature,421, 957.
Lou, Z., et al. (2006). MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Molecular Cell,21, 187–200.
Luo, K., Zhang, H., Wang, L., Yuan, J., & Lou, Z. (2012). Sumoylation of MDC1 is important for proper DNA damage response. EMBO Journal,31, 3008–3019.
Luo, K., et al. (2015). CDK-mediated RNF4 phosphorylation regulates homologous recombination in S-phase. Nucleic Acids Research,43, 5465–5475.
Luo, K., et al. (2016). A phosphorylation–deubiquitination cascade regulates the BRCA2–RAD51 axis in homologous recombination. Genes & Development,30, 2581–2595.
Ma, J., et al. (2012). PALB2 interacts with KEAP1 to promote NRF2 nuclear accumulation and function. Molecular and Cellular Biology,32, 1506–1517.
Ma, T., et al. (2013). RNF111-dependent neddylation activates DNA damage-induced ubiquitination. Molecular Cell,49, 897–907.
Machida, Y., Kim, M. S., & Machida, Y. J. (2012). Spartan/C1orf124 is important to prevent UV-induced mutagenesis. Cell Cycle,11, 3395–3402.
Mailand, N., et al. (2007). RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell,131, 887–900.
Mallette, F. A., et al. (2012). RNF8-and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. The EMBO Journal,31, 1865–1878.
Mattiroli, F., et al. (2012). RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell,150, 1182–1195.
Meredith, J., Jr., Fazeli, B., & Schwartz, M. (1993). The extracellular matrix as a cell survival factor. Molecular Biology of the Cell,4, 953–961.
Mosbech, A., et al. (2012). DVC1 (C1orf124) is a DNA damage–targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nature Structural & Molecular Biology,19, 1084.
Muñoz, M. C., et al. (2012). Ring finger nuclear factor RNF168 Is important for defects in homologous recombination caused by loss of the breast cancer susceptibility factor BRCA1. Journal of Biological Chemistry,287, 40618–40628.
Nakada, S., et al. (2010). Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature,466, 941.
Nijman, S. M., et al. (2005). A genomic and functional inventory of deubiquitinating enzymes. Cell,123, 773–786.
Nowsheen, S., et al. (2018a). ZNF506-dependent positive feedback loop regulates H2AX signaling after DNA damage. Nature Communications,9, 2736.
Nowsheen, S., et al. (2018b). L3MBTL2 orchestrates ubiquitin signalling by dictating the sequential recruitment of RNF8 and RNF168 after DNA damage. Nature Cell Biology,20, 455.
Orthwein, A., et al. (2014). Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science,344, 189–193.
Orthwein, A., et al. (2015). A mechanism for the suppression of homologous recombination in G1 cells. Nature,528, 422.
Pfeiffer, A., et al. (2017). Ataxin-3 consolidates the MDC1-dependent DNA double-strand break response by counteracting the SUMO-targeted ubiquitin ligase RNF4. EMBO Journal,36, 1066–1083.
Postow, L., & Funabiki, H. (2013). An SCF complex containing Fbxl12 mediates DNA damage-induced Ku80 ubiquitylation. Cell Cycle,12, 587–595.
Poulsen, M., Lukas, C., Lukas, J., Bekker-Jensen, S., & Mailand, N. (2012). Human RNF169 is a negative regulator of the ubiquitin-dependent response to DNA double-strand breaks. Journal of Cell Biology,197, 189–199.
Qin, B., et al. (2019). UFL1 promotes histone H4 ufmylation and ATM activation. Nature Communications,10, 1242.
Rabinovich, E., Kerem, A., Fröhlich, K.-U., Diamant, N., & Bar-Nun, S. (2002). AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Molecular and Cellular Biology,22, 626–634.
Ritchie, K. J., et al. (2004). Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nature Medicine,10, 1374.
Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S., & Bonner, W. M. (1998). DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. Journal of Biological Chemistry,273, 5858–5868.
Scheffner, M., Nuber, U., & Huibregtse, J. M. (1995). Protein ubiquitination involving an E1–E2–E3 enzyme ubiquitin thioester cascade. Nature,373, 81.
Schmidt, C. K., et al. (2015). Systematic E2 screening reveals a UBE2D–RNF138–CtIP axis promoting DNA repair. Nature Cell Biology,17, 1458.
Shao, G., et al. (2009). The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proceedings of the National Academy of Sciences,106, 3166–3171.
Shiloh, Y. (2003). ATM and related protein kinases: Safeguarding genome integrity. Nature Reviews Cancer,3, 155.
Skaug, B., & Chen, Z. J. (2010). Emerging role of ISG15 in antiviral immunity. Cell,143, 187–190.
Sobhian, B., et al. (2007). RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science,316, 1198–1202.
Stewart, G. S., et al. (2009). The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell,136, 420–434.
Stingele, J., Habermann, B., & Jentsch, S. (2015). DNA–protein crosslink repair: Proteases as DNA repair enzymes. Trends in Biochemical Sciences,40, 67–71.
Stingele, J., Schwarz, M. S., Bloemeke, N., Wolf, P. G., & Jentsch, S. (2014). A DNA-dependent protease involved in DNA–protein crosslink repair. Cell,158, 327–338.
Stingele, J., et al. (2016). Mechanism and regulation of DNA-protein crosslink repair by the DNA-dependent metalloprotease SPRTN. Molecular Cell,64, 688–703.
Sy, S. M., Huen, M. S., & Chen, J. (2009). PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proceedings of the National Academy of Sciences,106, 7155–7160.
Sy, S. M. H., Jiang, J., O, W. S., Deng, Y., & Huen, M. S. Y. (2013). The ubiquitin specific protease USP34 promotes ubiquitin signaling at DNA double-strand breaks. Nucleic Acids Research,41, 8572–8580.
Thorslund, T., et al. (2015). Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature,527, 389.
Typas, D., et al. (2016). The de-ubiquitylating enzymes USP26 and USP37 regulate homologous recombination by counteracting RAP80. Nucleic Acids Research,44, 2976.
Wang, B., & Elledge, S. J. (2007). Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proceedings of the National Academy of Sciences,104, 20759–20763.
Watanabe, K., et al. (2009). RAD18 promotes DNA double-strand break repair during G1 phase through chromatin retention of 53BP1. Nucleic Acids Research,37, 2176–2193.
Watanabe, S., et al. (2013). JMJD1C demethylates MDC1 to regulate the RNF8 and BRCA1–mediated chromatin response to DNA breaks. Nature Structural & Molecular Biology,20, 1425–1433.
Wei, Y., & Xu, X. (2016). UFMylation: A unique & fashionable modification for life. Genomics, Proteomics & Bioinformatics,14, 140–146.
Wijnhoven, P., et al. (2015). USP4 auto-deubiquitylation promotes homologous recombination. Molecular Cell,60, 362–373.
Wu, W., Koike, A., Takeshita, T., & Ohta, T. (2008). The ubiquitin E3 ligase activity of BRCA1 and its biological functions. Cell Division,3, 1–1.
Xiao, A., et al. (2009). WSTF regulates the H2A. X DNA damage response via a novel tyrosine kinase activity. Nature,457, 57.
Yau, R. G., et al. (2017). Assembly and function of heterotypic ubiquitin chains in cell-cycle and protein quality control. Cell,171, 918–933.e920.
Yin, Y., et al. (2012). SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes & Development,26, 1196–1208.
Zhang, Z., Vuori, K., Reed, J. C., & Ruoslahti, E. (1995). The alpha 5 beta 1 integrin supports survival of cells on fibronectin and up-regulates Bcl-2 expression. Proceedings of the National Academy of Sciences,92, 6161–6165.
Zhang, H., et al. (2016). A cell cycle-dependent BRCA1–UHRF1 cascade regulates DNA double-strand break repair pathway choice. Nature Communications,7, 10201.
Zhang, L., et al. (2018). RNF126 quenches RNF168 function in the DNA Damage response. Genomics, Proteomics & Bioinformatics,16, 428–438.
Zhong, W., Feng, H., Santiago, F. E., & Kipreos, E. T. (2003). CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature,423, 885.
Acknowledgements
This work was supported by funding from the National Institutes of Health (CA217183 and CA203561) to Zhenkun Lou.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nowsheen, S., Deng, M. & Lou, Z. Ubiquitin and the DNA double-strand break repair pathway. GENOME INSTAB. DIS. 1, 69–80 (2020). https://doi.org/10.1007/s42764-019-00007-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42764-019-00007-5