Abstract
Purpose
The deep learning-aided automatic bone age assessment systems have obtained the best results but still depending on the advanced methods to attain highly accurate region of interest (ROI) segmentation and error-free bone age assessment based on different regions of bone. By avoiding these drawbacks, this paper presents a comparative analysis on a deep learning-aided bone age assessment using 5 regions over 13 regions under Tanner-Whitehouse 3 (TW3) method.
Methods
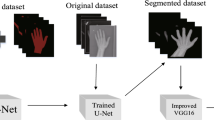
The segmentation of both 5 regions and 13 regions is employed by both adaptive Otsu thresholding and improved U-Net segmentation. Further, the adoption of enhanced “deep convolutional neural network (CNN) is used for the bone age assessment (BAA)” using the 13 region and 5 regions. The development of Opposition Searched Harris Hawks Optimization (OS-HHO) is preferable for both segmentation and prediction.
Results
From the analysis, the root mean square error (RMSE) of 5th region is 8.45% better than that of the 13th region. Similarly, the mean absolute squared error (MASE) of 5th region was 2.30% higher than the 13th region.
Conclusion
The assessment of bone age by 5 regions was better when compared with the 13 regions for the suggested bone age assessment (BAA) model, respectively.




















Similar content being viewed by others
References
Abbasi MU, Rashad A, Basalamah A, and Tariq M, Detection of epilepsy seizures in neo-natal EEG using LSTM architecture, IEEE Access, 2019; 7.
Alcina M, Lucea A, Salicru M, and Turbon D, Reliability of the Greulich & Pyle method for bone age estimation in a Spanish sample, J Forensic Leg Invest Sci. 2015; 1(3).
Antonio T, Juan A. A radius and ulna TW3 bone age assessment system. IEEE Trans Biomed Eng. 2008;55(5):1463–76.
Bakthula R and Agarwal S, Automated human bone age assessment using image processing methods – survey. Int J Comput Appl. 2014; 104(13).
Bojja GR, Ofori M, Liu J, Ambati LS (2020) Early public outlook on the coronavirus disease (COVID-19): a social media study. Social Media Analysis on Coronavirus (COVID-19).
Cao F, Huang H, Pietka E, Gilsanz V. Digital hand atlas and web-based bone age assessment: system design and implementation. Computerized Med Imag Graph. 2000;24:297–307.
Chemtex RM, Kantheti S. Kantheti S Classification of skin cancer using deep learning, convolutional neural networks -opportunities and vulnerabilities-a systematic review. Int J Modern Trends Sci Technol. 2020;6(11):101–8.
Chen X, Li J, Zhang Y, Lu Y, Liu S. Automatic feature extraction in X-ray image based on deep learning approach for determination of bone age. Future Gener Comput Syst. 2020;110:795–801.
Chen Y, Yang J, Qian J. Recurrent neural network for facial landmark detection. Neurocomputing. 2017;219:26–38.
Christoforidis A, Badouraki M, Katzos G, Athanassiou-Metaxa M. Bone age estimation and prediction of final height in patients with β-thalassaemia major: a comparison between the two most common methods. Pediatric Radiol. 2007;37:1241–6.
Deshmukh S and Khaparde A, Faster region-convolutional neural network oriented feature learning with optimal trained recurrent neural network for bone age assessment for pediatrics, Biomed. Signal Process. Control. 2022; 71.
Fischer B, Welter P, Gunther RW, Deserno TM. Web-based bone age assessment by content-based image retrieval for case-based reasoning. Int Jof Comput Assisted Radiol and Surgery. 2012;7(3):389–99.
Gertych A, Zhang A, Sayre J, Pospiech-Kurkowska S, Huang H. Bone age assessment of children using a digital hand atlas. Comput Med Imag Graph. 2007;31:322–31.
He J and Jiang D, Fully automatic model based on SE-ResNet for bone age assessment, in IEEE Access 2021; 9: 62460–62466.
Heidari AA, Mirjalili S, Faris H, Aljarah I, Mafarja M, Chen H. Harris hawks optimization: algorithm and applications. Future Gener Comput Syst. 2019;97:849–72.
Hussien AG et al 2020 Crow search algorithm: theory, recent advances, and applications, in IEEE Access, 2020; 8: 173548–173565.
Iglovikov V, Rakhlin A, Kalinin A, and Shvets A, Pediatric bone age assessment using deep convolutional neural networks, arXiv:1712.05053 2017.
Jagadeeshwar L. Tabjula, Srijith Kanakambaran, Sheetal Kalyani, Prabhu Rajagopal, Balaji Srinivasan " Outlier analysis for defect detection using sparse sampling in guided wave structural health monitoring," Structural control and Health Monitering, Volume 28, Issue, March 2021.
Jagadeeshwar Tabjula, S. Kalyani, Prabhu Rajagopal, Balaji Srinivasan "Statistics-based baseline-free approach for rapid inspection of delamination in composite structures using ultrasonic guided waves," Structural Health Monitoring, 2021.
Kim J, Shim W, Yoon H, Hong S, Lee J, Cho Y, Kim S. Computerized bone age estimation using deep learning based program: evaluation of accuracy and efficiency. Am J Roentgenol. 2017;209:1374–80.
King D, et al. Reproducibility of bone ages when performed by radiology registrars: an audit of Tanner and Whitehouse II versus Greulich and Pyle methods. The Brit J Radiol. 1994;67:848–51.
Kowdiki M, and Khaparde A, Automatic hand gesture recognition using hybrid meta-heuristic-based feature selection and classification with Dynamic Time Warping, Comput. Sci. Rev. 2021; 39.
Lee H, Tajmir S, Lee J, Zissen M, Yeshiwas BA, Alkasab TK, Choy G, Do S. Fully automated deep learning system for bone age assessment. J Digit Imaging. 2017;30:427–41.
Li K. et al., Automatic bone age assessment of adolescents based on weakly-supervised deep convolutional neural networks, in IEEE Access, 2021; 9: 120078–120087.
Liu J, Qi J, Liu Z, Ning Q, Luo X. Automatic bone age assessment based on intelligent algorithms and comparison with TW3 method. Comput Med Imag Graph. 2008;32:678–84.
Liu Y, Zhang C, Cheng J, Chen X, Wang ZJ. A multi-scale data fusion framework for bone age assessment with convolutional neural networks. Comput Biol Med. 2019;108:161–73.
Malipatil S, Maheshwari V and Chandra MB, Area optimization of CMOS full adder design using 3T XOR, Int. Conf.Wireless Commun. Signal Processing and Networking (WiSPNET), 2020; 192–194.
Mansourvar M, Ismail MA, Herawan T, Raj RG, Kareem SA, and Nasaruddin FH, Automated bone age assessment: motivation, taxonomies, and challenges, Comput. Math. Methods Med. 2013; 2013.
Markus H, Benedikt F, Hauke S, Thomas S. Support vector machine classification based on correlation prototypes applied to bone age assessment. IEEE J Biomed Health Inf. 2013;17(1):190–7.
Merzban MH, Elbayoumi M. Efficient solution of Otsu multilevel image thresholding: a comparative study. Expert Syst Appl. 2019;116:299–309.
Mirjalili S, Mirjalili SM, Lewis A. Grey wolf optimizer. Adv Eng Software. 2014;69:46–61.
Pan X, Zhao Y, Chen H, Wei D, Zhao C, Wei Z. Fully automated bone age assessment on large-scale hand X-ray dataset. Int J Biomed Imag. 2020;2020:1–12.
Pietka E, Gertych A, Pospiech S, Cao F, Huang H, Gilsanz V. Computer-assisted bone age assessment: image preprocessing and epiphyseal/ metaphyseal ROI extraction. IEEE Trans Med Imag. 2001;20(8):715–29.
Preetha NSN, Brammya G, Ramya R, and Praveena S, Grey wolf optimization-based feature selection and classification for facial emotion recognition, IET Biom. 2018; 7(5).
Ramesh S, Vydeki D, Recognition and classification of paddy leaf diseases using optimized deep neural network with Jaya algorithm, Inf. Process. in Agric. 2019.
Ren X, et al. Regression Convolutional neural network for automated pediatric bone age assessment from hand radiograph. IEEE J Biomed Health Inf. 2019;23(5):2030–8.
Ronneberger O, Fischer P, and Brox T, U-Net: convolutional networks for biomedical image segmentation, Springer: 2015; 234–241.
Son SJ. et al., TW3-based fully automated bone age assessment system using deep neural networks, in IEEE Access. 2019; 7: 33346–33358.
Spampinato C, Palazzo S, Giordano D, Aldinucci M, Leonardi R. Deep learning for automated skeletal bone age assessment in X-ray images. Med Image Anal. 2017;36:41–51.
Thodberg HH, Kreiborg S, Juul A, Pedersen KD. The BoneXpert method for automated determination of skeletal maturity. IEEE Trans Med Imag. 2009;28(1):52–66.
Wang D, Tan D, Liu L. Particle swarm optimization algorithm: an overview. Soft Comput. 2018;22(2):387–408.
Wibisono A & Mursanto P, Multi region-based feature connected layer (RB-FCL) of deep learning models for bone age assessment, J Big Data. 2020; 7.
Zarie M, Jahedsaravani A, Massinaei M Flotation froth image classification using convolutional neural networks, Miner Eng. 2020;155.
Acknowledgements
I would like to express my very great appreciation to the co-authors of this manuscript for their valuable and constructive suggestions during the planning and development of this research work.
Author information
Authors and Affiliations
Contributions
All authors have made substantial contributions to conception and design, revising the manuscript, and the final approval of the version to be published. Also, all authors agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding author
Ethics declarations
Ethics approval
Not applicable
Informed consent
Not applicable
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Deshmukh, S., Khaparde, A. Comparative analysis of 5 regions over 13 regions bone age assessment via TW3 method with deep learning. Res. Biomed. Eng. 38, 871–900 (2022). https://doi.org/10.1007/s42600-022-00225-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42600-022-00225-z