Abstract
In this paper, we present a retina abnormality classification framework for diabetic retinopathy and age related macular degeneration using content based image retrieval. This is performed in two phases, namely, feature extraction and pattern recognition. In the first phase, image pre-processing and Otsu multi-level thresholding is applied to retina fundus images to extract eleven low level spatial and statistical features. The second phase consists of machine learning based classification with these features using four machine learning classifiers, namely, Naive Bayes classifier, support vector machine, K-nearest neighbour classifier and random forest classifier. It is found that random forest classifier outperforms all the other classifiers for the detection of both bright and dark lesion classes and achieves 94.8% and 95.1% accuracy, respectively with ROC area 0.977 and 0.98, respectively.
Similar content being viewed by others
1 Introduction
Case based reasoning (CBR) systems [1,2,3] have gained popularity in recent times in Health Sciences as an aid to diagnosis, classification and treatment planning. In the medical domain, CBR shows promise as an appropriate method for diagnosis and therapy planning by using the symptoms given. However, in some CBR systems, feature extraction, feature mining and indexing are done prior to case formulation.The retrieval of images from case databases is essential for designing a CBR system. Computer Aided Diagnosis (CAD) systems using medical images perform extraction of relevant information including feature extraction to generate a diagnosis either automatically or semi automatically. CBR using Content Based Image Retrieval (CBIR) [8] has been used to manage digital images in medical databases. The retrieval process requires image processing steps in order to extract low level features from the image. Typically, CBR system is used to retrieve relevant cases from a case data base by finding the similarity of the test case and the prototype cases [13].This similarity is central to CBR [12] as the CBIR system banks on it. The problem space is regarded as a vector space and the case is represented as a feature vector. The similarity between the feature vectors of the test case and prototype cases is considered. For cases with heterogeneous structures in the medical domain, such as images and associated metadata, machine learning techniques are integrated with CBR [21, 22]. Furthermore, due to the increase in computing power in recent years, some emergent measures, like Random Forests [9] (RF) have been suggested in the work of Cunningham [12], for computing similarity.
Due to the threat of vision loss, DR diagnosis with the help of automated techniques has become an active area of research. DR in the earlier stages is regarded as Non-Proliferative Diabetic Retinopathy (NPDR), whereas, in the later stages, it becomes Proliferative Diabetic Retinopathy (PDR). The symptoms of NPDR that show up in retina fundus images can be classified as bright lesions and dark lesions based on the color feature. In the first stage of DR, the deformation of retinal capillary results in very small red spots called microaneurysms (MA). In the next stage, hard exudates (HE) are formed due to lipid formation from the thinned blood vessels. The prognosis of NPDR includes cotton wool spots (CWS) which are micro infracts caused by obstructed blood vessels. Haemorrhages (HAM) are formed when blood vessels leak blood into retina, with further progression of the disease, and are a major symptom in the advanced stage of NPDR and also for PDR. Due to the lack of oxygen supply in retina, new vessels are formed, which threaten the patient’s eye sight. MA and HAM are bright red and dark red in color, respectively and are considered as dark lesion classes whereas HE and CWS are yellow and whitish yellow, respectively, and are treated as bright lesion classes. Age related Macular Degeneration (AMD) is another threatening eye disease as it leads to permanent blindness of aged people, if not treated at an early stage. Macula lutea which is responsible for central vision gets degenerated for the formation of debris like structures [11] near it. This abnormal formation is named as drusen and the stage of the disease is called dry AMD. Drusens are yellow in color and circular in shape. Based on appearance, drusens are quite similar to HE of DR and are included in bright lesion classes. A CBR for retrieval using decision trees [5] have been explored earlier. RF as a classifier has also been studied [25, 30].
The present paper describes the approach based on image processing and RF classifier for abnormality detection of retina fundus images and also compares this technique with Support Vector Machine (SVM), K-nearest neighbour (KNN) and Naive Bayes (NB) classifiers. Machine learning using a probabilistic model was selected first, as decision making using probability is a common technique for the physicians. So, Naive Bayes classifier was selected. Then to increase the accuracy, SVM classifier was selected, as it avoids over-fitting. Once the boundary of different clusters is determined, a small amount of noisy data cannot affect the accuracy of the classifier much. When SVM is compared with Naive Bayes classifier, SVM achieved higher accuracy than Naive Bayes classifier for dark lesion classification [24]. RF classifier is selected because according to the literature survey, RF classifier produces remarkably accurate results. It is robust to larger databases [25]. Machine learning using Single Layer Perceptron (SLP) algorithm has also been tested and its performance is compared with SVM and NB classifier [31]. It was concluded that SLP and SVM give comparable results and both outperform NB classifier. KNN classifier is selected due to its simplicity to handle multi-class classification and robustness to noisy data. However, RF classifier is found to perform best.
The rest of the article is presented as follows: Sect. 2 describes the literature review on the related topic, Sect. 3 represents the algorithm and the methodology developed in this research work. In Sects. 4 and 5, the experimental results and the discussion on the results are given, respectively.The article is concluded in Sect. 6.
2 Related work
CBR based approaches are being implemented widely in health sciences [3]. According to the survey of Begum et al. [3], cultivation of CBR in medical sciences is of growing interest but it is prototyped for most of the systems. The mechanism of a CBR based approach is described broadly in the pioneering work of Aamodt and Plaza [1]. The survey, by Dasgupta and Banerjee [13], on different approaches using the combination of CBIR and CBR for therapy planning has been reported. The first process of any CBR based approach is retrieval of similar cases from case base when facing a new case. This retrieval is made on the basis of similarity measures, between the case base prototypes and test cases. Different methods for evaluating similarity measures are organized into a coherent taxonomy in the technical report of Cunningham [12]. Though it is argued that different similarity measure techniques might be compatible for different problems, the centrality of the representation of feature values due to direct similarity measures have been suggested by Cunningham [12]. A CBR based approach is developed [21] for medical case retrieval. More complex formats of information like videos [22] are included in the retrieval framework for processing incomplete information. The research work of Quellec et al. [22] is based on image processing as matching of images found in query documents and reference documents of the patient’s information is evaluated. The automatic detection of MA, the first clinical signs of DR, are developed in another research work of Quellec et al. [23]. The approach of forming an ensemble model with tree predictors are described in details in the pioneering work of Breiman [9]. In an earlier work [5], a CBIR framework using decision trees is used. The use of RF for lesion classification in DR is also described [25]. Another pioneering work in the detection of retina abnormalities due to DR and AMD is done by Niemeijer et al. [18, 19]. Bright lesion detection [18] is performed by machine learning. The machine learning algorithm is based on calculating the lesion-probability map which is used to identify whether a pixel is a part of a lesion or not. The ground truth of the training and test images are evaluated by the approval of human experts. For dark lesion detection [19], mathematical morphology based operations are used. A set of features were selected for classification of red lesions. According to the work of Osareh et al. [20], detection of retinal exudates are performed by using Fuzzy C means clustering followed by feature extraction and classification. The classification of objects is done using standard back propagation and scaled conjugate gradient descent approaches [20]. Current research of Akram et al. [2] suggested a combination of m-Mediods based Modelling and Gaussian Mixture Model forming a hybrid classifier to detect retina abnormalities. In their work [2], HE and CWS were clubbed together as exudates. A survey on the development of CAD system for the detection of ocular diseases [26] show that most of the research is focused on the detection of the presence of exudates due to DR, not the grading of the disease. As DR and AMD are diagnosed with similar signatures, it is necessary to identify and distinguish these two diseases clearly. RF classifier is used for improving ill-focused images [27] by extracting the useful information from spatial and transform domain. This information consists of features to train the trees of RF algorithm. RF classifier is used for the detection and diagnosis of lymphatic diseases [28] with Particle Swarm Optimization for feature selection. The original dataset with its re-sampling are used for training purpose of RF classifier. In the work done by Kar et. al. [4], DR in advanced stage called proliferative stage (PDR) or neovascularisation is considered and the development of new abnormal blood vessels, is detected. But in our current research work, the different signs of non-proliferative stages of DR like MA, HE and the starting signs of AMD are identified along with some proliferative stage abnormalities of DR like HAM. As early detection of abnormality can stop the growth of the diseases with medication, non-proliferative symptom detection is more important. In one of our earlier works [5], a decision tree was applied on a set of images for separating them to different classes. But no feature selection was done. In the current research work, we extend the framework on a larger database of retina images with feature selection and different machine learning techniques and also use RF classifier which uses an ensemble of decision trees. In the work of Safi et. al. [6], a survey on the previous works related to subclinical retinopathy biomarkers is carried out and summarized but no new approach for the detection of DR is proposed. In the work done by Shree et. al. [7], an image processing approach is developed for segmenting and analysing optic disk from colored retina image but detection of DR was not considered. The article of Acton et. al. [39] described a method for evidence based diagnosis of retinal images affected by AMD and Stargardt disease. A feature extraction method for improving the image retrieval process is proposed in this article [39]. The accuracy of the extraction of Stargardt disease images from the database used in the research work of [39] is 63.5% which is better than the traditional system. In the work of Yin et. al. [40], a decision generating system is developed depending on CBR for distinguishing the headache as migraine pain and tension-related pain. The recall and precision rates of the proposed system is higher than those of the guideline based system. The p-value of migraine pain and tension-related pain identification are 0.019 and 0.002 which are promising. In the article of Khussainova et. al. [41], a combination of CBR model with clustering is proposed for the better selection of cases in the retrieve step of CBR paradigm. The existing cases of CBR model are clustered into eight groups using K-means clustering. This significant step optimises the choices of the cases in the retrieval step and the average isocentre distances of the retrieved cases with the new case is decreased from 207.87 to 31.26 mm. For the diagnosis of cancer, the research work of Saraiva et. al. [42] proposed a CBR-based method combined with rule based reasoning and the system considers the patient’s clinical signs and personal parametric measures as the input and produces the type of cancer as a result. The proposed method in [42] improves the diagnosis efficiency with a p-value of 0.02664 which is promising. A knowledge support system based on CBR model is proposed in the work of Tyagi et. al. [43] for the risk prediction of the asthmatic patients. The concept of CBR model is used for the similarity measurement of new case to past cases.
3 Proposed methodology
A composite database of two hundred and ten images containing different stages of DR, dry AMD and normal retina is considered as input dataset, which is obtained by collecting the images from DIARETDB0 [16], DIARETDB1 [15], HRF database [10], e-ophtha [14], MESSIDOR [17]. In Table 1, the number of images collected from different standard databases are given.
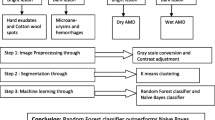
The geometric resolutions of the retina images from DIARETDB0, DIARETDB1, HRF database, e-ophtha and MESSIDOR are 1500 \(\times\) 1152, 1500 \(\times\) 1152, 3584 \(\times\) 2438, 2544 \(\times\) 1696 and 1440 \(\times\) 960, respectively with 24 bit depth each except MESSIDOR (bit depth = 8), and all these images are resized to 150 \(\times\) 115. These images contain hard exudates (HE), cotton wool spots (CWS) due to DR and drusens due to AMD as bright lesion classes and hemorrhages (HAM) and microaneurysms (MA) due to DR as dark lesion classes. In Fig. 1, the diagrammatic representation of the system is given.
3.1 Lesion detection
The color fundus images of retina with abnormalities like DR and AMD are taken as input. Then, green channel extraction followed by contrast enhancement is done, as green component reflects the best contrast suitable for segmentation.
Contrast enhancement Contrast enhancement is applied to the input images for improving the visual representation of those images. It is done by mapping the gray scale range of the input image to the full range [0 255]. The lowest gray scale value (low_in) is mapped to 0 and the highest gray scale value (high_in) is mapped to 255. The intermediate values (inter) are mapped using the function
Segmentation and extraction of region of interest (ROI) After contrast enhancement, Otsu multilevel segmentation is applied on the image. For segmentation, the number of clusters is considered as four. It is observed that mainly three different shades of color are present in retinal image: dark red for blood vessel tree and dark lesions like HAM; bright yellow for optic disc and bright lesions like exudates; reddish yellow for the fluid and membrane of retina. Black color indicates the corner portions of the fundus image. After segmentation, the region of interest (ROI) is extracted. For dark lesion detection, the cluster corresponding to darkest region is considered as the ROI. This includes dark lesions along with blood vessel tree. In the case of bright lesion detection, the cluster corresponding to brightest region is the ROI. In this case, the optic disc along with bright lesions is extracted. The next step is to eliminate the normal objects of retina from ROI, namely elimination of the optic disc and blood vessel tree.
Optic disc elimination Optic disc removal is an important step in the characterisation of bright lesions [7]. In the image containing bright clusters, optic disc is eliminated following the two steps outlined as follows: as the size of bright lesions is significantly small compared to the size of optic disc, lesions are removed from the image by the morphological closing operation. This leaves a circular structure covering the optic disc. Then image subtraction is done between the image containing bright lesions with optic disc and the image containing only optic disc. In Fig. 2, the different steps of lesion detection are shown. Figure 2a, b represents the input retina image and the contrast enhanced image. Figure 2c is the segmented image with four clusters. Figure 2d represents the image containing bright lesions and optic disc. After removing the small structures from Fig. 2d, a circular structure in the position of optic disc is obtained. Then, after removing the optic disc, lesions are detected in Fig. 2e. This retains the bright lesions only as output.
Blood vessel tress elimination In case of blood vessel tree elimination, image opening is used with disc type structuring element. This eliminates the dark lesions. Then upon applying logical XOR operation between the image containing dark lesion with blood vessel tree and the image containing only blood vessel tree, the blood vessel tree is removed. The input retina image, segmented image and the final output image is shown in Fig. 2. The lesion detection step using image processing implemented using Matlab can be applied to the databases containing macula centric retina fundus images without any manual adjustments.
a Input retina image with HE and CWS (image014 of DiaretDB1 [15]) b contrast enhanced image c segmented image by Otsu multilevel thresholding d extraction of the cluster containing bright pixels e detection of bright lesions
3.2 Feature selection
In addition to the choice of better data sets for accuracy of classification, designing of good features is also of paramount importance. Eleven features are selected for constructing the CBIR system using decision tree model. The selection of features is done depending on the consultation with the ophthalmologists and on literature review. Texture-based features are: color, shape, size and sharpness of edge. Color feature is important for distinguishing between dark lesions from bright lesions. For differentiation among bright lesion classes, color feature is also useful as HE is generally bright yellow and CWS is generally whitish yellow. In dark lesion classes, MA is normally light red in contrast to the dark red color of HAM. The variation of shape and size of MA compared to HAM is also significant. Sharpness of edge is selected as it contributes significant information about the edge of CWS which is blurred and the edge of HE which is sharp [18]. In Table 2, the texture based low level physical attributes of different classes of lesions are listed along with their categorical feature values. In the feature selection step, the evaluation of texture based features like color, shape, size and sharpness of edge of the abnormal objects needs manual interpretation. All the selected features are treated as extracted features for the evaluation of feature vector set as elimination of a single feature decreases the accuracy of the proposed methodology.
Area and perimeter in terms of pixel count are important as these evaluate the size and shape of the lesions in numeric values. Circularity is the measure of roundness of the lesions and works as a key feature to discriminate drusens from HE. Standard deviation and smoothness are two statistical features which gives significant information for distinguishing normal retina from the affected one. Eccentricity and Euler number is also an important evaluators of shape of an object and hence selected. Table 3 describes all the features selected in this research work and all these features presented in Table 3 are evaluated using Matlab code. A decision tree based approach (Figs. 3, 4 and 5) using J48 classifier of Weka [29] has been used in the feature selection process.
Each input retina image is evaluated to find the abnormalities and if any lesion is detected, then the selected feature values of that lesion are evaluated. In one retina image, several signs of DR and AMD may appear. All are detected and labelled by the Matlab function regionprop. Then with the help of the ground truth available with the standard databases mentioned in Table 1, the artefacts are identified. Each lesion contributes a feature vector with its class type. Thus a set of feature vectors is formed from all the input images. In this research work, a total of two hundred and ten images are considered as input image set which contributes three hundred and eleven feature vectors. This set is tested with 20 fold cross validation. So the whole set of feature vectors is subdivided into 20 subsamples. Among these 20 subsamples, 19 subsamples containing two hundred and ninety five set elements are considered as training set. On this training set, RF classifier is used and compared with SVM, KNN and NB classifiers. Detection of lesions using image processing steps is implemented using Matlab. The machine learning step using four classifiers is implemented using Weka [29] data mining tool.
Decision tree for feature selection (Here, cws = cotton wool spot, cir = circular, nu = not uniform, circu = circularity, he = hard exudates, sd = standard deviation, nor = normal, ma= microaneurysms, ham = haemorrhages, dry = dry AMD, eccen = eccentricity, euler = Euler number, vs = very small, med = medium)
Random forest classifier Random Forest classifier is an ensemble classifier which consists of a collection of several decision trees. Votes of all these decision trees make the final prediction of the class for a particular instance. In this classifier, trees are made deep to recognize different patterns. For training purposes, tree bagging is used. If \(X=(x_1, x_2,\ldots , x_n)\) be the data points with response variable \(Y=(y_1, y_2,\ldots ,y_n)\), then a decision tree f(t) is trained on \(x_t\), \(y_t\) where \(t \in\) {1,2,...,T}, \(x_t\) and \(y_t\) are sample of n training data. T is total number of trees in the forest. The prediction of the test set Z is made by majority vote. The total dataset is divided into two groups: two thirds of the data in this dataset are treated as training set and the rest of the data as the test set. The training set helps in the growth of the tree. Cases are randomly selected allowing replacement, that is, the case once selected for a growing a tree can be reconsidered for another tree. A total of m (=\(\sqrt{M}\) ) number of features are extracted randomly from the selected M number of features.The number of extracted features is kept fixed during the growth of the forest. The best split on these extracted features is used to divide a parent node of a tree. The test dataset is called Out-Of-Bag (OOB) data. The OOB data calculates the error rate of the classifier internally. For each test data, each tree votes for a class for each test case. The class with majority votes is assigned to the test case. In random forest classification, a number of decision tree classifiers are applied to different subsets of the dataset to be classified. The accuracy and over-fitting of the classifier is controlled by averaging of the outcomes from different sub-trees. Bagging decreases the variance of the classifier. If the trees are correlated, then the influence of noise in one tree affects the other trees also. By using bagging, de-correlation among the trees can be achieved by assigning them to different training set. In case of RF classifier, optimization involves feature bagging for each candidate tree. For optimization of RF classifier, number of features used for the training purpose at the each split of node is \(\sqrt{M}\) where M is the total number of features and the total number of trees used for voting is set to 100 in Weka .
Algorithm of random forest classifier
-
Step 1: Assignment of total number of classes to L and total number of features to M.
-
Step 2: Calculation of number of extracted features (m) (generally \(m=\sqrt{M}\) ).
-
Step 3: Random selection of a sub-dataset containing L different classes for each tree.
-
Step 4: Random selection of m features for the calculation of best split and decision for each node of a decision tree.
Support vector machine classifier SVM tries to fit a hyperplane to separate two datasets having different set of feature values. The criterion of fitting the hyperplane is to maximize the distance between the nearest data point of any of the class and the hyperplane itself.
Let T be the training data set containing n points: \((\overrightarrow{x_1} , y_1 ), (\overrightarrow{x_2}, y_2 ),\ldots , (\overrightarrow{x_n}, y_n)\). Each \(\overrightarrow{x_i }\) represents the data point and \(y_i\) has the value of either +1 or -1. Now the hyperplane must divide the set of data points in such a way that \(y_i\) with value +1 are separated from \(y_i\) with value -1. The hyperplane can be mathematically expressed as the set of points \(\overrightarrow{x}\) which satisfies the equation
where \(\overrightarrow{w}\) called normal vector. The value of \(\frac{b}{\Vert \overrightarrow{w}\Vert }\) is used for calculating the offset of the hyper-plane from the origin. A margin is the separator line to the closest training points.
The regularization parameter is used for specifying the margin of misclassification of the training data. A large value of the regularization parameter directs the optimizer to choose a smaller margin hyperplane which can correctly classify all the data points. Conversely, a small value of the regularization parameter tells the optimizer to select a larger margin hyperplane even if some of the data points may be misclassified by it. The gamma parameter evaluates the degree of influence of a training example. A low value of gamma parameter indicates that the points far away from the separating hyperplane are considered for the selection of the separating hyperplane. A high gamma value indicates the contrary. In Weka, we have used both Radial Basis Kernel (RBF) kernel and PolyKernel for classifying our dataset. The classification results produced by PolyKernel are better than that of RBF kernel. Along with that, PolyKernel is efficient in higher dimension and is less time consuming. Hence it is selected as kernel trick for SVM classifier.
Algorithm for support vector machine classifier
-
Step 1: For linearly separable data points, if p is the dimension of the data point vector, then (p-1) dimensional hyper-plane is identified which maximizes the margin of two classes.
-
Step 2: If the data points are not linearly separable, then non-linear hyper-plane is selected using the Kernel trick. A suitable kernel function is selected to solve the particular problem from several standard kernels.
K-nearest neighbor classifier The KNN algorithm assigns a new case to its class on the basis of the similarity of the new case with its neighbours. The neighbours are previously assigned to their representative classes. Two steps, namely, selection step and fusion step are followed in order, in KNN classification. In the selection step, evaluation of the similarity of a new case is done with the cases of the training set. Then, depending on the similarity measure, K number of most similar cases from the training set is selected. In the fusion step, the new case is assigned to the most frequent class among its K neighbours.
Generally, one optimization parameter called number of neighbours is selected as the square root of number of training instances. In this research work, among three hundred and eleven instances, two hundred and ninety five instances are used as training data and hence the value of K is set to seventeen here. Another optimization parameter of KNN classifier is distance calculation which is set as Euclidian distance function in Weka.
Algorithm for KNN classifier
-
Let (\(F_i\), \(C_i\)) be the cases where \(F_i\) denotes the the \(i^{th}\) feature vector and \(C_i\) denotes its class. If M be the new case to be assigned to a class, the following steps are to be executed:
-
Step 1: Evaluation of distances (generally by Euclidian distance function) between M and \(F_i\) where (i=1,2,...,n).
-
Step 2: Sorting of the distances in non-decreasing order.
-
Step 3: Selection of top K distances from the sorted list, where K is the number of neighbors selected by the classifier.
-
Step 4: Selection of K points corresponding to the specified K neighbors.
-
Step 5: Selection of the class voted by majority of these K neighbors.
-
Step 6: Assignment of M to that class.
Naive Bayes classifier Naive Bayes classifier is based on the theory of conditional probability with strong independence assumption between the attributes. The class value of an instance is calculated using the conditional probabilities of a set of features. If \(A_1\), \(A_2\),..., \(A_n\) are considered as independent attributes and C is the class value, then according to Bayes theorem,
For an instance, all the class values are calculated and the class with highest probability is assigned to the instance. For Naive Bayes classifier, 20 fold cross validation is used for optimization.
Algorithm for Naive Bayes classifier
-
Step 1: Let S be the set of feature vectors and each element of S contains m attribute values (m is number of selected features).
-
Step 2: Let X = (\(x_1\), \(x_2\),..., \(x_m\)) be the new feature vector to be classified.
-
Step 3: Let p be the number of different classes i.e. \(C_1, C_2,\ldots , C_p\).
-
Step 4: X is assigned to the class \(C_i\) iff \(P(C_i|X) > P (C_j|X)\) for \(1 \le j \le p, j \ne i\) and \(P(C_i|X)\) is evaluated using Eq. (2).
4 Results
The performance analysis of all four classifiers considered in this paper is carried out using the confusion matrix in Weka [29] machine learning classifier. For each classifier’s confusion matrix, number of True Positive, True Negative, False Positive and False Negative instances of each class are evaluated. These values are further used to calculate sensitivity, specificity, accuracy, F-measure and MCC. The measures used for evaluating the efficiency of the classifiers are described in Eqs. (4)–(7).
-
\(TP_i\) (True Positive): number of instances correctly predicted as of class \(X_i\)
-
\(TN_i\) (True Negative): number of instances predicted as not of class \(X_i\) and actually not of class \(X_i\)
-
\(FP_i\) (False Positive): number of instances incorrectly predicted as of class \(X_i\)
-
\(FN_i\) (False Negative): number of instances predicted as not of class \(X_i\) but actually of class \(X_i\)
-
Accuracy (Acc): the efficiency of the classifier to correctly classify a member
$$\begin{aligned} Acc_i=\frac{TP_i+TN_i}{TP_i+TN_i+FP_i+FN_i} \end{aligned}$$(4) -
Sensitivity (Sn) : efficiency to detect the positive class
$$\begin{aligned} Sn_i= \frac{TP_i}{TP_i+FN_i} \end{aligned}$$(5) -
Specificity (Sp): efficiency to detect the negative class
$$\begin{aligned} Sp_i=\frac{TN_i}{TN_i+FP_i} \end{aligned}$$(6) -
Mathew Correlation Coefficients (MCC): estimate of over prediction and under prediction is defined as
$$\begin{aligned} MCC_i=\frac{(TP_i \times TN_i) - (FP_i \times FN_i)}{\sqrt{TP_i+ FP_i) (TP_i + FN_i)(TN_i+FP_i) (TN_i+FN_i}} \end{aligned}$$(7)
MCC generates three different types of values: \(-1\), 0 and 1. The value \(-1\) means the classifier’s prediction is incorrect, 0 means random prediction and 1 means fully correct prediction. Tables 4, 5, 6 and 7 describes the performance measures of RF, SVM, KNN and NB classifier, respectively.
Tables 8 and 9 show some interesting facts about the classifiers’ performances on our dataset using 20 fold cross validation. RF and SVM classifiers are better for bright lesion classes compared to other two classifiers. RF classifier achieves more accuracy with a coverage of more ROC area for classifying dark lesion classes. The ROC area achieved by SVM is also very close to KNN classifiers. KNN classifier is a close competitor to SVM classifier in terms of accuracy and ROC area.
5 Discussion
Recently, CBR, a special paradigm based on machine learning that emulates a physician’s diagnosis process based on similar cases in the past, has become popular in diagnosis and therapy planning in a variety of applications [37]. In order to implement such a process on the computer, a case database must be developed that contains instances of the diseases to be diagnosed. Our work is an attempt to develop this database for diagnosis of DR and AMD, which are metabolic diseases of the retina that can lead to blindness unless timely detected. In order to aid the physician in accurate diagnosis, the signatures of the disease must be correctly extracted and identified. For this purpose, image pre-processing and segmentation using Otsu multilevel segmentation are carried out.Then the feature selection process must be carried out to input these features into a machine learning classifier. We have used low level features like color, texture and other statistical features as shown in Tables 2 and 3. A decision tree based approach has been used to accurately quantify these features as shown in Fig. 3. The reason for selecting decision tree based model is its easy interpretability. The decision tree model evaluates the classifiers’ performance on the training set with different subsets of features and finally the subset, which performs best, is selected. The decision tree has been expanded in Figs. 4 and 5 to further quantify two other features, Euler number and area, respectively. For classification of the multiple lesions of DR, namely, MA, HE, CWS and HAM and drusens for AMD, four classifiers, RF, SVM, KNN and NB have been used, and their performances are given in Tables 4, 5, 6 and 7, in terms of sensitivity, specificity, accuracy, MCC, ROC area and F measure. It is found that RF and SVM give better results than KNN and NB. RF also outperforms SVM, and has thus been chosen in designing the case database. In our earlier works, we have used NB, SVM and RF classifiers for machine learning [30, 31]. In the current paper, we have included KNN classifier.
RF has several advantages as described below:
-
As for computational complexity, RF classifier outperforms SVM because, if number of data points is n in the set, then n \(\times\) n matrix is generated by SVM in one of its intermediate step which increases its computational complexity.
-
RF classifier is better suited for large number of training data points than SVM as over fitting is a problem of SVM.
-
RF classifier is also robust to noisy data.
-
Furthermore, as MA is the first clinical signs of DR, the RF classifier can detect it with higher accuracy, adding an advantage for choosing RF classifier. So, RF classifier can be treated as the best option for classification of bright and dark lesions due to DR and AMD with the selected feature set.
Along with that, RF classifier has been found to outperform the other three and is thus chosen. The advantages of RF over the other three classifiers are described in Table 10.
A bar chart depicting accuracy of the four classifiers for bright and dark lesions is given in Fig. 6.
Several groups have reported the development of case database for CBIR [21, 22, 32,33,34,35]. Quellec et. al. [21] have reported multimedia case retrieval of retina images, using decision tree and by fusing heterogeneous information in [22]. Quellec et. al. [33] have coupled Bayesian networks with Dezert–Smarandache theory [36] for multimodal case retrieval from retinal images. Elsayed et. al. [34] have reported a CBR system for image categorisation using time series analysis and Dynamic Time Wrapping (DTW). Pilot studies using digital photographic analysis to extract low level features of HE and MA for DR and drusen, have been conducted by Chaum et. al. [35]. The work of Hijazi et. al [38] proposed a time series analysis based approach for the classification of AMD in retina images. The reported sensitivities have been compared with other work in Table 11.
In contrast to other methods, which are confined to one or two lesions, we have considered multiple lesions that exist during the earlier stages of DR and AMD. The novelty of this current paper lies in the combination of the machine learning approach and the concept of CBIR approach to develop a CBR system for early detection of DR and AMD. Also, a combination of well-known segmentation algorithms and well-established machine learning algorithms is performed to provide an efficient CBR based method.
Also, a combination of well-known segmentation algorithms combined with well-established machine learning algorithms to provide an efficient CBR database for efficient CBIR using suitable features has been attempted in this paper.
6 Conclusion
This paper is an attempt to develop a CBIR system for developing CBR approach to the detection of retina abnormalities caused by DR and AMD. DR and AMD are two leading causes of vision loss all over the world. This research is an effort to assist the doctors for the timely and accurate detection of the earliest signs of these two diseases.The early detection of these diseases can suppress the progress of the diseases and minimize the loss of vision. The database includes retina fundus images with lesions due to DR and AMD. The images are collected from standard databases like DiaretDB0 [16], DiaretDB1 [15]. The approach of a physician for the treatment of a patient mostly depends on his experiences and knowledge base. Retina fundus images of case base are processed through segmentation using Otsu multilevel thresholding and then detection of abnormal objects are performed after eliminating optic disc and blood vessel tree. Eleven features (listed in Table 3) based on physical appearance and texture of the detected abnormal regions is selected. A set of feature vectors formed by the evaluation of features for each abnormal object is split into training and test set for machine learning. Four classifiers have been used: RF, SVM, KNN and NB. It was found that RF was best suited for our work. From the analysis of performance of the above mentioned four classifiers, it is observed that with the selected feature set and case base, RF classifier achieves 94.8% accuracy for predicting bright lesion classes with 0.977 as ROC AUC (Area Under Curve). This result is best among all four classifiers in terms of accuracy and very close in terms of ROC AUC to that of the other three classifiers. RF classifier performs better than the others for the prediction of dark lesion classes. The accuracy and ROC AUC of RF classifier for detecting dark lesion classes is 95.1% and 0.98, respectively. It may be mentioned here that, some other classifiers have also been tested but the performance of these was not satisfactory compared to these four classifiers. As our target of our problem is to classify an object, regression techniques like linear and logistic regression are abandoned. The well-known classification techniques like decision tree, neural network, SLP [24], SVM, NB, KNN and RF classifiers are considered. Approaches like single and multi-layer perceptron based on neural network and the approach of RF classifier based on decision tree are both capable of producing high accuracy. As no single approach produces best results for all types of cases, it is better to compare their performances on a chosen dataset. With the data sets considered in our study, it was found that RF outperforms all other classifiers considered above.
Future work will also consist of including more images in the case database to provide greater accuracy, as well as using images from local hospitals to further upgrade the case database. Hence CBIR using CBR database applying RF classifier is a promising candidate for a CAD system for diagnosis of retina abnormalities.
References
Aamodt A, Plaza E (1994) Case-based reasoning: foundational issues, methodological variations, and system approaches. AI Commun 7(1):39–59
Akram MU, Khalid S, Tariq A, Khan SA, Azam F (2014) Detection and classification of retinal lesions for grading of diabetic retinopathy. Comput Biol Med 45:161–171
Begum S, Ahmed MU, Funk P, Xiong N, Folke M (2011) Case-based reasoning systems in health sciences: a survey of recent trends and developments. IEEE Trans Syst Man Cybern Part C (Appl Rev) 41(4):421–434
Kar S, Maity S (2017) Detection of neovascularization in retinal images using mutual information maximization. Comput Electr Eng 62:01–15
Banerjee S, RoyChowdhury A (2015) Case based reasoning in the detection of retinal abnormalities using decision trees. Proc Comput Sci 46:402–408
Safi H, Safi S, Hafezi-Moghadam A, Ahmadieh H (2018) Early detection of diabetic retinopathy. Surv Ophthalmol. https://doi.org/10.1016/j.survophthal.2018.04.003
Shree TDV, Revanth K, Sri Madhava Raja N, Rajinikanth V (2018) A hybrid image processing approach to examine abnormality in retinal optic disc. Proce Comput Sci 125:157–164
Smeulders AWM, Worring M, Santini S, Gupta A, Jain R (2000) Content-based image retrieval at the end of the early years. IEEE Trans Pattern Anal Mach Intell 22(12):1349–1380
Breiman L (2001) Random forests. Mach Learn 45(1):05–32
Budai A, Bock R, Maier A, Hornegger J, Michelson G (2013) Robust vessel segmentation in fundus images. Int J Biomed Imaging 2013:01–11
Crabb JW, Miyagi M, Gu X, Shadrach K, West KA, Sakaguchi H, Kamei M, Hasan A, Yan L, Rayborn ME, Salomon RG, Hollyfield JG (2002) Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc Natl Acad Sci 99(23):14682–14687
Cunningham P (2008) A taxonomy of similarity mechanisms for case-based reasoning. Technical report UCD-CSI 2008-01, University College Dublin
Dasgupta M, Banerjee S (2014) Similarity based retrieval in case based reasoning for analysis of medical images. World Acad Sci Eng Technol Int J Comput Electr Autom Control Inf Eng 8(3):539–545
Decencière E et al (2013) TeleOphta: machine learning and image processing methods for teleophthalmology. IRBM. https://doi.org/10.1016/j.irbm.2013.01.010
Kauppi T, Kalesnykiene V, Kamarainen JK, Lensu L, Sorri I, Raninen A, Voutilainen R, Uusitalo H, Kälviäinen H, Pietilä J (2007) DIARETDB1: diabetic retinopathy database and evaluation protocol, Technical report
Kauppi T, Kalesnykiene V, Kamarainen JK, Lensu L, Sorri I, Uusitalo H, Kalviainen H, Pietila J (2006) DIARETDB0: Evaluation database and methodology for diabetic retinopathy algorithms, Technical report
Decencière et al. (2014) Feedback on a publicly distributed database: the Messidor database. Image Anal Stereol 33(3):231–234 ISSN 1854–5165. http://www.iasiss.org/ojs/IAS/article/view/1155 or http://dx.doi.org/10.5566/ias.1155
Niemeijer M, Ginneken BV, Russell SR, Suttorp-Schulten MSA, Abramoff MD (2007) Automated detection and differentiation of drusen, exudates, and Cotton–Wool spots in digital color fundus photographs for diabetic retinopathy diagnosis. Investig Ophthalmol Vis Sci 48(5):2260–2267
Niemeijer M, Ginneken BV, Staal J, Suttorp-Schulten MSA, Abramoff MD (2005) Automatic detection of red lesions in digital color fundus photographs. IEEE Trans Med Imaging 24(5):584–592
Osareh A, Mirmehdi M, Thomas B, Markham R (2003) Automated identification of diabetic retinal exudates in digital colour images. Br J Ophthalmol 87:1220–1223
Quellec G, Lamard M, Bekri L, Cazuguel G, Cochener B, Roux C (2007) Multimedia medical case retrieval using decision trees. Conf Proc IEEE Eng Med Biol Soc 1:4536–4539. https://doi.org/10.1109/IEMBS.2007.4353348
Quellec G, Lamard M, Cazuguel G, Roux C, Cochener B (2011) Case retrieval in medical databases by fusing heterogeneous information. IEEE Trans Med Imaging Instt Electr Electron Eng 30(1):108–118
Quellec G, Lamard M, Josselin PM, Cazuguel G, Cochener B, Roux C (2008) Optimal wavelet transform for the detection of microaneurysms in retina photographs. IEEE Trans Med Imaging 27(9):1230–1241
Saha R, Roychowdhury A, Banerjee S (2016) Diabetic retinopathy related lesions detection and classification using machine learning technology. Artif Intell Soft Comput (ICAISC) 9693:734–745 Springer International Publishing, Switzerland
Roychowdhury A, Banerjee S (2017) Random forests in the classification of diabetic retinopathy retinal images, advanced computational and communication paradigms. In: Proceedings of international conference on advanced computational and communication paradigms (ICACCP - 2017) 1:168-176
Zhang Z, Srivastava R, Liu H, Chen X, Duan L, Kee Wong DW, Kwoh CK, Wong TY, Liu J (2014) A survey on computer aided diagnosis for ocular diseases. BMC Med Inf Dec Making 14(80):01–29. https://doi.org/10.1186/1472-6947-14-80
Kausar N, Majid A (2016) Random forest based scheme using feature and decision levels information for multi-focus image fusion. Pattern Anal Appl 19(1):221–236 Springer
Almayyan W (2016) Lymph diseases prediction using random forest and particle swarm optimization. J Intell Learn Syst Appl 8:51–62
Witten IH, Frank E, Hall M (2011) Data mining: practical machine learning tools and techniques, 3rd edn. Morgan Kaufmann, Burlington, Massachusetts
Roychowdhury A, Chatterjee T, Banerjee S (2018) A random forest classifier-based approach in the detection of abnormalities in the retina. Med Biol Eng Comput 57(1):193–203. https://doi.org/10.1007/s11517-018-1878-0
Saha R, Roychowdhury A, Banerjee S, Chatterjee T (2018) Detection of retinal abnormalities using machine learning methodologies. Neural Netw World 28:457–471. https://doi.org/10.14311/NNW.2018.28.025
Lamard M, Cazuguel G, Quellec G, Bekri L, Roux C, Cochener B (2007) Content based image retrieval based on wavelet transform coefficients distribution. Conf Proc IEEE Eng Med Biol Soc 1:4532–4535. https://doi.org/10.1109/IEMBS.2007.4353347
Quellec G, Lamard M, Cazuguel G, Roux C, Cochener B (2008) Multimodal medical case retrieval using the Dezert–Smarandache theory. Conf Proc IEEE Eng Med Biol Soc 1:394–397. https://doi.org/10.1109/IEMBS.2008.4649173
Elsayed A, Hijazi MHAH, Coenen F, Garcia-Finana M, Sluming V, Zheng Y (2011) Image categorization using time series case based reasoning. International conference on case based reasoning (ICCBR (2011) Lecture notes in computer science. Springer, Berlin, p 6880. https://doi.org/10.1007/978-3-642-23291-6_31
Chaum E, Karnowski TP, Govindasamy VP, Abdelrahman M, Tobin KW (2008) Automated diagnosis of retinopathy by content-based image retrieval. Retian J Retin Vitr Dis 8(10):1463–1477. https://doi.org/10.1097/IAE.0b013e31818356dd
Smarandache F, Dezert J (2006) Advances and applications of DSmT for information fusion. American Research Press, Rohoboth
Choudhury N, Begum SA (2016) A survey on case-based reasoning in medicine. Int J Adv Comput Sci Appl 7(8):136–144
Hijazi MHA, Coenen F, Zheng Y (2010) Retinal image classification using a histogram based approach In: International joint conference on neural networks (IJCNN 2010). https://doi.org/10.1109/IJCNN.2010.5596320
Acton ST, Soliz P, Russell S, Pattichis MS (2008) Content bBased image retrieval: the foundation for future case-based and evidence-based ophthalmology. In: IEEE international conference on multimedia and expo 1:541–544. https://doi.org/10.1109/ICME.2008.4607491
Yin Z, Dong Z, Lu X, Yu S, Chen X, Duan H (2015) A clinical decision support system for the diagnosis of probable migraine and probable tension-type headache based on case-based reasoning. J Headache Pain 16:29. https://doi.org/10.1186/s10194-015-0512-x
Khussainova G, Petrovic S, Jagannathan R (2015) Retrieval with clustering in a case-based reasoning system for radiotherapy treatment planning, mini EURO conference on im-proving healthcare: new challenges, new approaches. J Phys Conf Ser 616:012013. https://doi.org/10.1088/1742-6596/616/1/012013
Saraiva RM, Bezerra J, Perkusich M, Almeida H, Siebra C (2015) A hybrid approach using case-based reasoning and rule-based reasoning to support cancer diagnosis: a pilot study. Stud Health Technol Inf 216:862–866
Tyagi A, Singh P (2015) ACS: asthma care services with the help of case base reasoning technique. Proc Comput Sci 48:561–567 Elsevier
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Roy Chowdhury, A., Banerjee, S. & Chatterjee, T. A cybernetic systems approach to abnormality detection in retina images using case based reasoning. SN Appl. Sci. 2, 1414 (2020). https://doi.org/10.1007/s42452-020-3187-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42452-020-3187-0