Abstract
Medical imaging describes the noninvasive methods that enable medical practitioners to look inside the body, which are critical in modern clinical diagnosis. The diagnosed medical images may have confidential information related to patient’s privacy. Privacy and security must be guaranteed for the digital images over Internet, to ensure its confidentiality. In this work, a novel encryption method is suggested for medical images. The work is intended to suggest a novel encryption scheme for medical images which is strong, secure and efficient. Also, it can be applied to any kind of the medical images, irrespective of its storage format. In the proposed method, the pixels of the image are shuffled using a pseudorandom number generator based on two-dimensional Zaslavski map. The permuted image is encrypted by DNA encryption. Various visual analysis, correlation analysis, quality and security analysis techniques are applied to verify the performance of the method.
Similar content being viewed by others
1 Introduction
Because of the rapid advancement of multimedia and communication technology, digital images are often used for communication. Medical images include radiography, computed tomography (CT), magnetic resonance imaging (MRI), photoacoustic imaging, etc. A patient’s medical image may include his medical records along with text-based personal information, clinical diagnosis and examination records [1]. With the recent developments in technology, medical field through E-health, or telemedicine is popular. In health sector, medical imaging has become a major part of most diagnostic procedures. Medical images can inform the doctor about the internal problems which could not be detected by external assessment of the patient. Unfortunately, traditional encryption algorithms such as DES, AES, etc., are not well suited for medical images due to their large size and different storage formats. Many encryption techniques exist to overcome the spying of information in Internet and to assure protection of patient’s private information.
Xiao Chen and Chun-Jie Hu suggested an ‘Adaptive medical image encryption algorithm based on multiple chaotic mapping’ [2]. In this work, multiple chaotic maps are applied in permutation phase and XOR operation for diffusion. Bhasker Mondal et al. describe a light weight secure encryption scheme based on chaos and DNA computing for grayscale images, in which encryption is implemented using 1D logistic map [3]. Dridi Manal and Mtibaa Abdellatif explain the encryption of medical images using DCT and two-dimensional Arnold Cat map in the paper ‘crypto-compression of medical image based on DCT and chaotic system’ [4].
In this work, we use an encryption algorithm which has two phases, pixel permutation using chaotic map and diffusion using DNA cryptography. Many of the recent works show that chaotic map is appropriate to generate pseudorandom sequences. Chaos-based cryptosystems are suitable for image encryption owing to the inherent properties of chaotic systems such as simple structure, ergodicity and high sensitivity to initial values and control parameters [5]. In the permutation phase, we have used a pseudorandom number generator based on two-dimensional Zaslavski map [6]. Diffusion phase makes use of DNA encryption in order to improve security of the system.
The rest of this paper is organized as follows. Section II describes the properties of chaotic system and the pros of 2D Zaslavski map. The encryption method is explained in Section III. The experimental results are shown in Section IV. Section V shows the performance analysis of the algorithm using various tests. Finally, conclusion is drawn in section VI.
2 Chaotic map in cryptography
Chaos is subtle nonlinear dynamical systems that are highly sensitive to initial conditions. The possibility for self-synchronization of chaotic oscillations has sparked an avalanche of works on application of chaos in cryptography [6]. The strong mixing property, sensitive dependence on initial conditions, continuous broadband power spectrum and control parameters make chaotic map advisable for cryptographic primitives. As chaotic maps are non-period, non-converging, discrete-time dynamical systems, they can be used to generate pseudorandom series. Also, because chaotic maps are cryptographically secure random number generators, they can be good enough to be used in image encryption.
2.1 2D Zaslavski map
In this work, two-dimensional Zaslavski map is used to create a pseudorandom number generator in the permutation phase. Sensitiveness to the control parameters makes this chaotic system very useful for many cryptographic applications which require pseudo-randomness [7]. A discrete-time dynamical Zaslavski map is defined by the following equation Eq. (1), where (x0, y0) is the initial condition. ϵ = 9.1,ʋ = 12.6695 and τ = 3.0 are system parameters, and ‘e’ is exponentiation.
Since Zaslavski map has a relevant randomness, less autocorrelation among elements and a high value for maximal Lyapunov exponent, it is used in the permutation phase. Randomness of Zaslavski map is analyzed by calculating maximal Lyapunov exponent and autocorrelation analysis. 2D Zaslavski map is found to have a positive Lyapunov exponent [8] and hence found to be suitable here. Figure 1 shows autocorrelation of pseudorandom elements generated by 2D Zaslavski map. The equality distribution of a chaotic sequence is good enough if its arithmetic mean is about to zero [9]. A pseudorandom number generator is implemented by applying the 2D Zaslavski map using Eq. (2), where Yi is the sequence generated by 2D Zaslavski map.
3 Methodology
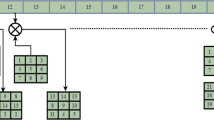
The proposed encryption scheme has two phases—permutation of pixel positions and diffusion of the permuted image. Figure 2 shows the steps in the proposed method. A pseudorandom series S is generated using the 2D Zaslavski map. The sequence S contains M x N of elements, where the number of pixels of the image is also M x N. The pixels of the image are rearranged as a matrix I of size (1, M x N). The elements of matrix I are reordered in the order of elements of the pseudorandom sequence S. The obtained matrix is constructed as the permuted image.
In the diffusion phase, we applied the principles of DNA encryption. DNA is a chain of nucleic acids: Adenine(A), Thymine(T), Guanine(G), Cytosine(C). A pixel of the image can be represented as the combination of four nucleic acids using DNA digital coding [10] patterns. It is possible to write eight DNA coding patterns, which satisfies the biological complementarity of bases.
Each pixel of the permuted image is represented with DNA patterns by randomly selecting one of the eight patterns. From the biomedical and genomic database NCBI, one of the considerably large DNA sequences is selected for key generation. NCBI is the major resource of bioinformatics tools and services. For our experimental simulation, a DNA sequence with more than 400,000 bases is accessed from the database. N bases from the accessed DNA sequence are taken randomly to form the key for encryption, since the DNA representation of the image also has N bases.
The DNA sequence which represents the image is added with the key DNA sequence using DNA addition operation. DNA sequences can be added and subtracted according to DNA addition and subtraction rules shown in Tables 1 and 2. DNA addition is analogous to binary addition. For example, if A = 00 and T = 11, then A + T=00 + 11 = 11. The DNA sequence obtained after addition operation is reconstructed as image which represents the encrypted image. Here, the encrypted image has same size and dimensions of the original image. In decryption, we are using DNA subtraction. DNA subtraction is according to the binary subtraction. The decrypted image has the same size of the original image.
4 Simulation result
Computed tomography(CT) is a valuable medical tool that helps a physician in medical diagnosis. Here, CT image of brain is used as plain image. The work is implemented in MATLAB 7. The original image, encrypted image and the decrypted image of the CT scan image of brain are shown in Fig. 3. From the results in the figure, it is clear that the cipher image is totally different from the plain image and also the encrypted image has the same size and dimension of the plain image. The method can be applied on images of any size and format.
5 Performance analysis
The performance of the proposed method is tested and analyzed with various metrics as shown below.
5.1 Randomness tests
The pseudorandom sequence generated is analyzed by randomness tests and considering autocorrelation of elements in the sequence. Autocorrelation exhibits the correlation among elements in the sequence. For random sequence, autocorrelation is preferred to be closer to zero at every time unit. Figure 4 shows the plot of autocorrelation of the elements in the pseudorandom sequence. Randomness of the pseudorandom sequence is tested by runs test, rank test, frequency monobit test, frequency test within a block, binary rank test and DFT test which are defined in NIST test suite [11]. The results of randomness test are given in Table 3. A positive value of P shows that the sequence passes the randomness tests that were done.
5.2 Security analysis
5.2.1 Histogram analysis
Histograms of image represent pixel intensity values in it. It is the graphical representation of tonal distribution of an image. Histogram analysis is widely used in image processing to analyze image quality and thereby comparing two images. Figures 5 and 6 show the result of histogram analysis.
Histogram of the encrypted image must be totally different from that of original image [12]. Since a good image encryption method tends to encrypt a plain image to random-like, it is desired to see a uniformly distributed histogram for a cipher image [12]. Hence, the method is found to be secure.
5.2.2 Histogram deviation
Histogram deviation [13] measures the deviation between histograms of original and encrypted images to ensure the quality of encryption algorithm. It is used as a common measure to analyze the quality of encryption methods in terms of how it maximizes the deviation between original and the cipher image [13]. Histogram deviation can be measured using Eq. 3.
where di is the amplitude of the absolute difference at the gray level i for an image with M X N pixels. A high value of DH in Table 4 shows high deviation between original and encrypted image. From the results, it is clear that the proposed method generates an encrypted image which is totally different from the plain image; hence, the method is secure.
5.2.3 Irregular deviation
Irregular deviation [13] measures how much deviation occurred in the image is irregular, because of encryption. Equations 4 and 5 show the calculation of irregular deviation.
where
where H(i) is the histogram of the absolute difference between original and encrypted image, MH is the mean value of histogram of the encrypted image and HD is the absolute of histogram deviations from its mean value. A preferable encryption method produces minimum irregular deviation. From the results shown in Table 4, it is clear that the proposed encryption method causes negligible irregular deviation, which assures security.
5.2.4 Deviation from ideality
Deviation from ideality explains how the encryption algorithm minimizes the deviation of the encrypted image from an assumed ideally encrypted image [13]. An ideally encrypted image CI must have a completely uniform histogram distribution, which means that the probability of existence of any gray level is constant [13] A low value for deviation from ideality ensures the performance of the encryption method. For a pixel in ideal encrypted image of M × N pixels,
elsewhere
where D represents the deviation in histogram of encrypted image C from the histogram of ideal encrypted image C1. Table 4 shows the obtained values of deviation from ideality. The obtained values are negligibly small so that the proposed method is found to be secure.
Table 4 shows the values of irregular deviation, histogram deviation and deviation from ideality obtained for various medical images when encrypted. The results show that the encrypted image has a high histogram deviation from the original image and negligible irregular deviation and minimum deviation from ideally encrypted image.
5.3 Correlation analysis
5.3.1 Correlation distribution
There exists a high correlation among neighboring pixels in an image. The high correlation among pixels in the original image is represented by the straight line it converges to, where in the encrypted image the correlation plot is scattered. Figures 7 and 8 show the correlation distribution of original and encrypted images, respectively.
From the obtained results, it can be concluded that the proposed encryption scheme minimizes the correlation among pixels in the plain image.
5.3.2 Correlation coefficient analysis
Correlation coefficient is a measure to analyze the similarity or coincidence between two images. The method is applied on different medical images, and the values of correlation coefficient are shown in Table 5. A high correlation coefficient represents high similarity. A correlation closer to zero shows minimum analogy between images. From the results, it is clear that the proposed method reduces the correlation between encrypted and plain images to a value closer to zero.
5.3.3 Pixel correlation
There exists a high correlation among neighboring pixels in an image. This helps in reconstruction of image if a portion of it is known. Encryption reduces correlation of the adjacent pixels of the image. Table 6 shows the correlation among adjacent horizontal, vertical and diagonal pixels in the image before and after encryption. The proposed method minimizes the correlation of nearby pixels which ensures the performance of the encryption scheme.
5.4 Analysis of differential attack
Analysis of differential attack is testing of how the change in information of the input affects the output. Differential attack is applicable primarily to block ciphers working on binary sequences [14], and it becomes a common attack to be considered in cipher design. The widely used methods of differential attack in image processing are analysis of NPCR and UACI metrics.
5.4.1 NPCR analysis
The number of pixel changing rate (NPCR) is one of the most common quantities used to evaluate the strength of image encryption algorithms with respect to differential attack [14]. NPCR represents the absolute number of pixel changing rate in the cipher image if one of the pixels of original image is modified. Equation 7 shows the calculation of NPCR. Higher the percentage of NPCR, stronger the encryption method will be. The proposed system results in NPCR closer to 100%, as included in Table 7, where C1 and C2 are the encrypted images of original images which differ in only one pixel value. From the results, we can conclude that even a very small change in the plain image generates a totally different cipher image.
5.4.2 UACI analysis
Unified averaged change in intensity (UACI) [14] is a metric used to test the effect of one pixel change in the whole image, encrypted by an encryption algorithm. It is widely used to analyze differential attack in image encryption. It focuses on the averaged difference between two encrypted images [14]. UACI can be defined as Eq. 8.
where C1 and C2 are two cipher images whose original images are differed in only a single pixel value. UACI obtained for the proposed method, shown in Table 7, is closer to the ideal value, which also ensures the robustness of the proposed scheme against differential attack.
5.5 Key space analysis
In the proposed method, the secret key includes the control parameters of 2D Zaslavski map as well as the DNA sequence taken from database of NCBI. The DNA sequence has the same number of bases in the DNA representation of the image pixels. Also, the encrypted image depends on the DNA digital coding pattern used in encryption. Figure 9 shows the result obtained when the image is decrypted with a key sequence with difference in one DNA base. Table 8 shows the results when the decrypted image is compared with the image decrypted with a DNA sequence with one base difference.
The results in Fig. 9 and Table 8 assure the key sensitivity and hence the security of the encryption scheme.
5.6 Quality analysis
5.6.1 Peak signal to noise ratio(PSNR)
PSNR indicates the changes in pixel values between the plain image and the cipher image [15]. A higher value of PSNR reflects image quality, and hence, the encrypted image will have a low PSNR. The preferred value of PSNR for an encrypted image is less than 10 dB [15]. Estimation of PSNR is shown in Eq. 9.
The obtained values of PSNR for various images are found to be less than 10 dB, which ensures the quality of the method.
5.6.2 Information entropy and SSIM
Information entropy [16] is a measure of randomness of an image. Information entropy of an image can be obtained by Eq. 10 in which Pk represents the probability distribution of the image. The value of entropy for an ideally random image is 8. The entropy of various cipher images calculated is listed in Table 9 which is very closer to the desirable value.
The loss of structural information due to encryption can be estimated by calculating SSIM(structural similarity in images) [15] of the cipher image. SSIM is used to compare the contrast luminance and structure of two images. The obtained entropy shows the randomness generated in the image after encryption. Also, the SSIM obtained ensures the dissimilarity between plain nd encrypted images.
5.6.3 Mean square error(MSE)
Mean square error (MSE) is the cumulative squared error between two digital images and can be used to check the avalanche effect [17]. Avalanche effect shows how a small change in the original image affects the cipher image. For two cipher images C1 and C2 which were encrypted with keys differ in one bit, the mean square error is calculated using Eq. 11. Value of MSE will be zero if both images are identical [17].
High value of MSE in Table 9 represents robustness to differential attack and thereby ensuring the quality of encryption scheme.
5.6.4 Image fidelity(IF) and structural content(SC)
Image fidelity [18] is a measure of accuracy, used to predict the image quality. A larger value of image fidelity represents poor quality of encryption algorithm. Eq. 12 shows the calculation of image fidelity.
where x and y are the original and cipher image, respectively. Structural content (SC) [18], a metric used to test the quality of image, can be obtained from Eq. 13, where x is the plain image and y is the encrypted image. A good encryption scheme has less value for structural content
The proposed method minimum information fidelity and structural content, from which the quality of the scheme is guaranteed.
The obtained values of PSNR, MSE, IF, SC, entropy and SSIM shown in Table 9 assure quality of the proposed method.
5.7 Mean value analysis
Mean value analysis is used for finding the vertical distribution of mean pixel gray values of an image [19]. The mean value varies along the width of a plain image. For an encrypted image, the mean value remains consistent along the width of the image, which is an indication of uniform distribution of values along the vertical lines of the encrypted image [19]. Results of mean value analysis in various images are shown im Fig. 10.
The proposed method is compared with some existing methods, and the results are shown in Tables 10 and 11. The values of NPCR, UACI, image entropy, correlation coefficients given in [4] and [5] are compared with the obtained values. The proposed method has a better result when compared with other existing methods.
6 Conclusion
The paper proposes a secure encryption method based on DNA cryptography and two-dimensional Zaslavski map for various medical images. There are two phases: permutation and diffusion. A detailed performance analysis is done to test, analyze and ensure the strength, security and quality of the method. Randomness of the pseudorandom sequence which is used in permutation is tested by various randomness tests and autocorrelation analysis. Visual analysis is done by various histogram analysis methods. High value of NPCR and UACI shows that the proposed encryption scheme is robust against differential attack. Apart from this, the quality of the encryption method is tested. The experimental results show that a slight variation in the key yields a highly uncorrelated image compared to the plain image. The results are found to be authentic, and hence, the method is well suited for medical image encryption.
7 Future scope
The method can be enhanced by encrypting the extracted features of the image other than the whole image. This can reduce time required for execution.
References
Zhou Y, Panetta K (2009) A lossless encryption method for medical images using edge maps. 31st annual ınternational conference of the IEEE EMBS, Minneapolis, Minnesota, USA, 2–6 September 2009
Chen X, Chun-Jie H (2017) Adaptive medical image encryption algorithm based on multiple chaotic mapping. Saudi J Biol Sci 24:1821–1827
Mondal B, Mandal T (2017) A light weight secure image encryption scheme based on chaos & DNA computing. J King Saud Univ Comput Inf Sci 29:499–504
Dridi M, Bouallegue B, Mtibaa A (2014) Crypto-compression of medical image Based on DCT and chaotic system. 2014 Glob Summit Comput Inf Technol (GSCIT). https://doi.org/10.1109/GSCIT.2014.6970113
Hanchinamani G, Kulakarni L (2014) Image encryption based on 2d Zaslavski map and pseudo hadmard transform. Int J Hybrid Inf Technol 7(4):185–200
Kocarev L (2001) Chaos-based cryptography: a brief overview. IEEE Circuits Syst Mag 1(3):8–21
Hamza R, Titouna F (2016) A novel sensitive image encryption algorithm based on the Zaslavsky chaotic map. Inf Secur J A Glob Perspect 21(4–6):162–179 ISSN: 1939–3555
Chirakkarottu S, Mathew S (2018) A comparative analysis of image encryption techniques using chaotic maps and DNA cryptography. J Adv Res Dyn Control Syst 10:301–314 15-Special Issue
Stoyanov B, Kordov K (2014) Novel Zaslavsky map based pseudorandom bit generation scheme. Appl Math Sci 8(178):8883–8887
Ramadan N, Ahmed HEH, Elkhamy SE, El-Samie FEA (2016) Chaos-based image encryption using an ımproved quadratic chaotic map. Am J Signal Process 6(1):1–13
El-Samie FEA, Ahmed HEH, Elashry IF, Shahieen MH, Faragallah O, El-Rabaie ESM, Alshebeili SA (2013) Image encryption, a communication perspective. Taylor & Francis group, Milton Park, pp 40–44
Wu Y, Yang G (2012) Image encryption using the two-dimensional logistic chaotic map. J Electron Imaging 21(1):013014
Ismail SM, Said LA, Radwan AG, Madian AH, Abu-Elyazeed MF (2018) Generalized double-humped logistic map-based medical image encryption. J Adv Res 10:85–98
Stoyanov B, Kordov K (2014) Novel zaslavsky map based pseudorandom bit generation scheme. Appl Math Sci 8(178):8883–8887
Ahmad J, Ahmed F (2010) Efficiency analysis and security evaluation of image encryption schemes. IJVIPNS-IJENS, pp 18–31
Hor A, Ziou D (2010) Image quality metrics: PSNR versus SSIM. 2010 ınternational conference on pattern recognition
Rajput Y, Gulve AK (2014) A comparative performance analysis of an image encryption technique using extended Hill Cipher. Int Journal of Comput Appl 95(4):17–20
Kumar R, Rattan M (2012) Analysis of various quality metrics for medical image processing. Int J Adv Res Comput Sci Softw Eng 2(11):137–144
Somaraj S, Hussain MA (2015) Performance and security analysis for image encryption using key image. Indian J Sci Tech 8(35):2–4. https://doi.org/10.17485/ijst/2015/v8i35/73141 ISSN (Print) : 0974–6846
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chirakkarottu, S., Mathew, S. A novel encryption method for medical images using 2D Zaslavski map and DNA cryptography. SN Appl. Sci. 2, 1 (2020). https://doi.org/10.1007/s42452-019-1685-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42452-019-1685-8