Abstract
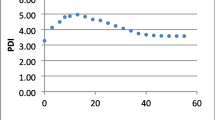
An experiment was conducted at the orchard of Chaudhary Charan Singh, Haryana, Agricultural University, Hisar (75°46′E and 29°10′N) and 215.2 a.m.s.l. The Ber powdery mildew index was recorded during the peak period (44–49 standard meteorological weeks) at different time intervals for the years 2015, 2016 and 2017 on Ber cultivar Umran. The study was carried out to find out the best fit growth models and significant weather variables in the disease progress curve. Five growth models viz. Exponential, Gompertz, Logistic, Log Logistic and Weibull were analysed for summarizing and comparing different plant disease epidemics. The statistical package R was used to illustrate models and disease progress over time. The best fit growth model was found by Akaike Information Criterion and Bayesian Information Criterion. The R code was developed and the regression analysis was performed to find out the best weather variables in the disease progress curve. It was observed that Exponential model was best fit growth model for the disease progress over time during 2015, whereas the Gompertz model was best fit during 2016 and 2017. The maximum and minimum temperature showed significant correlation with powdery mildew during 2015, whereas during 2017 the powdery mildew has significantly and negatively correlation with minimum temperature (− 0.74) and evening relative humidity (− 0.72). The coefficient of determination (R2) during 2015 and 2017 was found 78 and 68%, respectively.



Similar content being viewed by others
References
Akaike H (1974) A new look at the statistical model identification. IEEE Trans Autom Control 19:716–723
Glawe DA (2008) The powdery mildews: a review of the world’s most familiar (yet poorly known) plant pathogens. Annu Rev Phytopathol 46:27–51
Gupta JH (1988) Perpetuation and epidemiology of powdery mildew of mango. Acta Hortic 231:528–533
Jamadar MM, Venkatesh H, Balikai RA, Patil DR (2009) Forecasting of powdery mildew disease incidence on Ber (ziziphus mauritiana lam.) based on weather. Acta Hortic 840:63
Jat RG, Goyal SK (2009) Epidemiological studies on powdery mildew of Ber, ziziphus mauritiana Lamarck. In: SHS Acta Hort I international jujube symposium, vol 840, pp 62
Jeger MJ (1984) The use of mathematical models in plant disease epidemiology. Sci Hortic 35:11–27
Kumar A, Solanki KR, Singh R (2000) Effect of rainfall on incidence of powdery mildew in ber. Ann Arid Zone 39(1):89–92
Mckinney HH (1923) Influences of soil temperature and moisture in inhibition of wheat seedling by Helminthosporium sativum. J Agric Res 26(2):171–179
Schwarz G (1978) Estimating the dimension of a model. Ann Stat 6:461–464
Sinha P, Prajneshu R, Varma A (2002) Growth models for powdery mildew development of mango. Ann Plant Prot Sci 10:84–87
Szabelska A, Siatkowski M, Goszczurna T, Zyprych J (2010) Comparison of growth models in package R. Nauka Przyroda Technol. 4(4):50
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
Fitting the growth model in R software
The following code shows how to analyze the data through R code
dataset1 < -read.csv(file.choose(),header = TRUE)
attach(dataset1)
result.m_1 < -drm(response ~ time, data = dataset1, fct = EXD.3())
summary(result.m_1)
plot(result.m_1)
result.m_2 < -drm(response ~ time, data = dataset1, fct = G.4())
summary(result.m_2)
plot(result.m_2)
result.m_3 < -drm(response ~ time, data = dataset1, fct = L.5())
summary(result.m_3)
plot(result.m_3)
result.m_4 < -drm(response ~ time, data = dataset1, fct = LL.5())
summary(result.m_4)
plot(result.m_4)
result.m_5 < -drm(response ~ time, data = dataset1, fct = W1.4())
summary(result.m_5)
plot(result.m_5)
AIC(result.m_1, result.m_2, result.m_3, result.m_4, result.m_5)
AIC(result.m_1, result.m_2, result.m_3, result.m_4, result.m_5,
+ k = Log(nrow(dataset1)))
BIC(result.m_1, result.m_2, result.m_3, result.m_4, result.m_5)
dataset2 < -read.csv(file.choose(),header = TRUE)
attach(dataset2)
result.m_1 < -drm(response ~ time, data = dataset2, fct = EXD.3())
summary(result.m_1)
result.m_2 < -drm(response ~ time, data = dataset2, fct = G.4())
summary(result.m_2)
result.m_3 < -drm(response ~ time, data = dataset2, fct = L.5())
summary(result.m_3)
result.m_4 < -drm(response ~ time, data = dataset2, fct = LL.5())
summary(result.m_4)
result.m_5 < -drm(response ~ time, data = dataset2, fct = W1.4())
summary(result.m_5)
AIC(result.m_1, result.m_2, result.m_3, result.m_4, result.m_5)
BIC(result.m_1, result.m_2, result.m_3, result.m_4, result.m_5)
dataset3 < -read.csv(file.choose(),header = TRUE)
attach(dataset3)
result.m_1 < -drm(response ~ time, data = dataset3, fct = EXD.3())
summary(result.m_1)
result.m_2 < -drm(response ~ time, data = dataset3, fct = G.4())
summary(result.m_2)
result.m_3 < -drm(response ~ time, data = dataset3, fct = L.5())
Rights and permissions
About this article
Cite this article
Singh, R., Kumar, M., Mamta, D. et al. Development of growth model for Ber powdery mildew in relation to weather parameters. Indian Phytopathology 72, 235–241 (2019). https://doi.org/10.1007/s42360-019-00124-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42360-019-00124-x