Abstract
Plum pox virus (PPV) a potyvirus in the family Potyviridae, is the etiologic agent of sharka, the worldwide most important and detrimental viral disease of stone fruits (genus Prunus). Although PPV was identified in Italy already in the 1970s and it has been largely reported in almost all Italian regions, a broad investigation and genetic characterization of PPV isolates was lacking in most of them, including Tuscany (Central Italy). To address this knowledge gap, in 2020 and 2021, field surveys were carried out in 80 commercial Tuscany orchards and a total of 690 leaf samples were collected from different Prunus species. PPV was identified in 25 and 12 peach and plum samples, respectively (more than 5% of tested samples), whereas no positive samples were reported in apricot. Eighteen of the 37 PPV positive samples showed mixed infections with other viruses and viroids, mostly Prunus necrotic ringspot virus in plum and peach latent mosaic viroid in peach. Molecular typing of PPV infected samples generated NIb/CP amplicons corresponding to PPV-Marcus (M) or PPV-Recombinant (Rec) strains. Furthermore, starting from the identification of eight PPV nucleotide sequences (among which five and two new PPV-M and PPV-Rec isolates, respectively), this study firstly identified the PPV-Mb subgroup in Italy, which was even prevalent than PPV-Ma. Finally, PPV-Rec isolates resulted phylogenetically close to Italian and Turkish isolates previously detected. Overall, the results here presented represent an important step to fill knowledge gaps about PPV in Tuscany, and we believe it may encourage other similar research to achieve more accurate data on PPV populations at both national and international levels.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plum pox virus (PPV), a potyvirus in the family Potyviridae, is the etiologic agent of sharka (also known as ‘plum pox’), the most important and detrimental viral disease of stone fruits (genus Prunus in the Rosaceae family). The main hosts of PPV are commercial cultivars and rootstocks of major Prunus species, such as apricot (P. armeniaca), peach (P. persica), plum (P. domestica), sweet (P. avium) and sour cherry (P.cerasus; García et al., 2014). In addition, PPV has been reported in several ornamental and wild Prunus species, such as blackthorn (P. spinosa) and cherry plum (P. cerasifera; James and Thompson, 2006; Sebestyén et al., 2008), as well as in two cases in Tilia sp. and Spirea sp. (Çıtır et al., 2018; Pigliónico et al., 2021). In Prunus species, PPV infection has been often associated with other viruses (e.g., apple chlorotic leaf spot virus, ACLSV, prune dwarf virus, PDV, Prunus necrotic ringspot virus, PNRSV) and viroids (e.g., hop stunt viroid, HSVd, peach latent mosaic viroid, PLMVd; Hadidi et al., 2011).
The PPV spread at local scale is mainly due to aphid species such as Myzus persicae, Aphis spiricola and Hyalopterus pruni, which transmit the virus in a non-persistent manner, whereas the long-distance spread of PPV has taken place through global trade of infected plant material (García et al., 2014). Identified for the first time in Bulgaria in 1918 on plum trees (Atanasoff, 1932), nowadays PPV is widespread in Europe, in the Mediterranean basin, in the Middle East, in North and South America, and in Central, South and East Asia (EPPO, 2022).
Sharka symptoms commonly occur on Prunus leaves and fruits as chlorotic (e.g., blotches, rings, oak-leaves patterns, vein clearing) and morphological alterations (e.g., distortion on leaves, arabesque depressions with necrotic areas on fruits; Revers and García, 2015). Therefore, although sharka poses no danger to consumers, it can make stone fruits unmarketable, also by causing acidity and deformities. Moreover, the only way to manage the disease is to destroy all infected tree. The impact of sharka disease over 30 years was thus estimated to be over 10,000 million euros, taking into account the economic losses in production and the costs of sanitary control and surveillance, and of eradication programmes run in affected countries worldwide (Cambra et al., 2006; Rubio et al., 2017). For these reasons, PPV was ranked as quarantine pest in European Union since 2000, although in 2019 it was regraded to regulated non-quarantine pest (RNQP) due to its widespread endemic presence (European Union, 2019).
With a filamentous flexible-shape capside of approximatively 750 × 15 nm (Sochor et al., 2012), the PPV genome consists of a positive sense single-strand RNA (+ ssRNA) of about 9,700-9,800 nucleotides (Schneider et al., 2011; Glasa et al., 2013). PPV sequencing allowed to detect the presence of one main ORF coding for a 355 kDa polyprotein (García et al., 2014). Polyprotein processing gave ten mature proteins renamed in the same way of the nucleotide sequences: P1, HCPro, P3, 6K1, CI, 6K2, VPg, NIapro, Nib, CP. There was also an additional protein, named P3N-PIPO, deriving from a short ORF embedded within the P3 encoding region (Revers and García, 2015). Among these proteins, P1, HCPro and Nlapro are viral proteases that operate for the polyprotein cleavage releasing proteins for virus replication and spreading (García et al., 2014). According to the well-known high mutation rates and high evolutionary potential of potyvirus (LaTourette et al., 2022), the worldwide PPV genetic characterization identified ten main strains: PPV-Marcus (M), PPV-Dideron (D), PPV-Recombinant (Rec), PPV-El Amar (EA), PPV-Cherry (C), PPV-Winona (W), PPV-Turkey (T), PPV-Cherry Russian (CR), PPV-Ancestor (An) and a recent PPV-Cherry Volga (CV; Chirkov et al., 2018). PPV-M, PPV-D and PPV-Rec have been mostly reported, with PPV-M resulting the most aggressive, commonly associated with severe disease symptoms and efficiently transmitted by aphids, whereas PPV-D being the most widely distributed, and PPV-Rec being the recombinant product of PPV-M with PPV-D. Conversely, the other strains are worldwide distributed but restricted to local areas (James et al., 2013).
Although PPV was identified in Italy already in the 1970s (Albert et al., 1974) and it has been largely reported in all Italian regions, except only for Umbria, Liguria and Aosta Valley, only three studies have focused on the genetic characterization of PPV Italian isolates (Myrta et al., 2005; Dallot et al., 2011; Rizza et al., 2014), identifying a few nucleotide sequences from Apulia, Sicily and other unknown areas (a few sequences from Emilia Romagna, Tuscany, Veneto and unknown areas exist as direct submissions to GenBank). In Tuscany, where PPV was firstly identified in the early 1990s (Ginanni et al., 1993), and which is one of the major regions of Central Italy for stone fruits production (ISTAT, 2021), a broad investigation and genetic characterization of PPV isolates was lacking. To address this knowledge gap, this study aimed to (i) confirm the presence of PPV in Tuscan stone fruit orchards, also detailing the rates of PPV co-infection with other viruses (i.e., ACLSV, PDV, PNRSV) and viroids (i.e., HSVd, PLMVd), and (ii) characterize at genetic level the obtained isolates. We anticipate that outcomes here reported will be useful for the understanding of genetic diversity and evolutionary relationships of PPV, thus supporting the challenging contrast to the detrimental sharka disease.
Materials and methods
Field surveys and sampling
In 2020 and 2021 (June-September), field surveys were carried out in 80 commercial orchards selected in accordance with the Regional Phytosanitary Service of Tuscany and located in the six Tuscan districts where almost all the production of stone fruits occur, i.e., Arezzo, Florence, Grosseto, Lucca, Pisa and Siena (Fig. 1). A total of 690 leaf samples were collected from different Prunus species, namely apricot (134), peach (336), and plum (220). These samples were randomly collected from different branches of tree crowns (only a few samples showed virus-like symptoms), placed in coolers and same-day carried to the Plant Pathology Lab of the Department of Agriculture, Food and Environment, University of Pisa where they were stored at 4 °C until RNA extraction.
Tuscany stone fruit orchards investigated for plum pox virus presence. Orchards with negative (white), and positive peach (grey), plum (black), and peach and plum (grey/black) samples are reported. Districts: AR, Arezzo; FI, Florence; GR, Grosseto; LI, Leghorn; LU, Lucca; MS, Massa-Carrara; PI, Pisa; PO, Prato; PT, Pistoia; SI, Siena
RNA isolation and cDNA synthesis
Cetyltrimethylammonium bromide buffer 2% (CTAB) was used to extract total nucleic acids (TNA) from 0.5 g of leaf tissue (Li et al., 2008) with minor modifications (Pedrelli et al., 2021). This tissue was powdered in liquid nitrogen and added to 5 ml 2% CTAB buffer and incubated at 65 °C for 15 min. TNA was extracted by one volume of chloroform:iso-amylalcohol (24:1) and precipitated with one volume of isopropanol. Pellets were then washed with 70% ethanol, air-dried and dissolved in 80 µl of RNase/DNase free water. RNA purification kit (EURx, Gdańsk, Poland) was employed, and samples were stored at -80 °C. cDNA synthesis was finally performed using M-MMLV reverse transcriptase (GeneSpin s.r.l., Milan, Italy), according to the manufacturers’ instructions.
PPV and other virus and viroid detection
Samples were first tested for the PPV presence by quantitative polymerase chain reaction (qPCR). Samples positive to PPV were then tested for co-infections with ACLSV, PDV, PNRSV, HSVd and PLMVd. Virus detections were achieved using protocols targeting coat protein gene (CP), according to Olmos et al. (2005) for PPV (PPV-D, PPV-M and PPV-Rec strains), Osman et al. (2017) for ACLSV, and Kim et al. (2010) for PDV and PNRSV. Viroid detections were obtained amplifying a small portion of HSVd and PLMVd genomes, according to Luigi and Faggioli (2011) and (2013), respectively. Infected and healthy controls were included as references in each assay. All samples were tested to determine the presence and quality of cDNA using 18 S rRNA as endogenous control (Osman et al., 2008). qPCR assays were conducted in 20 µl reaction volume contained ITaq (Bio-Rad Laboratories, Hercules, CA, USA) using a Rotor-Gene Q Thermocycler (Qiagen, Venlo, The Netherlands).
PPV sequencing and in silico assays
Molecular typing of PPV strains (i.e., PPV-D, PPV-M and PPV-Rec) was achieved in a C-1000 touch Thermocycler (Bio-Rad Laboratories) using primers spanning nuclear inclusion bodies and coat protein (NIb/CP) region (Subr et al., 2004). Infected and healthy controls were included as references. The PCR products directly sequenced by Sanger DNA method (Eurofins genomics, Ebersberg, Germany) were compared using BLASTn program in GenBank with available isolates (www.ncbi.nlm.nih.gov) and sequence alignments were carried out in Bioedit (Hall, 1999). The phylogenetic trees were obtained in Mega X (Kumar et al., 2018), using Maximum Likelihood (ML) method (Jukes-Cantor model) and bootstrap replicated 1,000 times. Available isolates containing partial or complete NIb/CP genomic region previously described in GenBank were employed in phylogenetic analysis and their accession numbers composing ad hoc database are reported in Supplementary Table S1. The PPV strains (C, D, EA, T, W) were used as out-groups.
Results and discussion
PPV is still present in Tuscany orchards, often as co-infection with other viruses and viroids
Despite the regulation that since 2000 obliged farmers to destroy orchards when resulting PPV infected, this study showed that PPV is still present in Tuscany, likewise in other Mediterranean and European areas (Yardimci and Culal-Klllc, 2011; Gospodaryk et al., 2013; Zagrai et al., 2022), supporting the recent downregulation to RNQP (European Union, 2019). Virus-like symptoms, i.e., chlorotic fathering and chlorotic ring on leaves, were observed in six peaches and six plums, respectively. The PCR assays reported PPV presence in 25 and 12 peach and plum samples, respectively (more than 5% of tested samples), whereas no positive samples were reported in apricot (internal control 18s RNA confirmed cDNA good quality with Ct values ranging between 10 and 14, data not shown). Of the symptomatic samples, only seven, i.e., one peach and six plums, were found to be infected. PPV positive samples were reported in Arezzo (n = 24), Florence (n = 12) and Lucca (n = 1) districts, whereas all samples from Grosseto, Pisa and Siena tested negative (Fig. 1).
Eighteen of the 37 PPV positive samples (49%) showed mixed infections. In most of these cases, PNRSV was the main co-infection (27%), followed by PLMVd (24%), whereas ACLSV and PDV were notably less reported (3% each), although some straight differences were observed between plum and peach. Only PNRSV was co-present (3 samples) among symptomatic samples. Moreover, PNRSV was the only virus co-detected in plum (6 samples), although previous studies showed lower PNSRV rates compared to other viruses (Salem et al., 2003; Çelik et al., 2022). The higher PNRSV occurrence observed in the present study could be due to different cultivar susceptibility (Borisova and Sotirov, 2021) and/or to the presence of PNRSV unalike isolates (Kinoti et al., 2017). Differently, PLMVd was the most co-detected in peach (9 samples), followed by PNRSV (4 samples) and then ACLSV and PDV (1 sample each). The higher occurrence of PLMVd in peach is in accordance with previous reports from Italian orchards (Faggioli and Barba, 2006), and this outcome may be due to the fact that PLMVd frequently occurs without leaf symptoms, making more difficult its management (Flores et al., 2006; Serra et al., 2017). Mixed infections of three viruses, namely PPV + ACLSV + PLMVd, PPV + PDV + PNRSV and PPV + PLMVd + PNRSV, were each reported only in one peach sample. Here, HSVd was never detected, despite its wide host range (Luigi and Faggioli, 2013) and its earlier reports in Tuscany on kumquat, mandarin-lime, florentine citron, pear and lemon (Ragozzino et al., 2005; Rizzo et al., 2017).
PPV-M strain was mostly reported in peach, PPV-Rec was only detected in plum
After almost 30 years from it first report (Ginanni et al., 1993), this is the first broad molecular characterization of PPV strains in Tuscany, as well as one of the few at the Italian regional level, after Myrta et al. (2005) in Apulia and Rizza et al. (2014) in Sicily. Molecular typing of PPV infected samples generated NIb/CP amplicon corresponding to PPV-M (459 bp) or PPV-Rec strains (605 bp), whereas PPV-D was never obtained. Actually, PPV-M was detected in all the 25 peach samples, but only in one plum sample, as PPV-Rec was found only in the remaining 11 plum samples. This divergence between species is in accordance with Sihelská et al. (2017) who reported different strains in different hosts (i.e., PPV-M in peach, PPV-Rec in plum, although also PPV-D in plum and apricot). No mixed strain infections were observed. The overall higher occurrence of PPV-M than PPV-Rec was likely because PPV-M is characterized by an efficient transmission in fields by aphids (James et al., 2013). This outcome is in accordance with García et al. (2014) describing PPV isolates pathogenicity as influenced by specific genetic features, as well as their epidemiological behaviour affected by host and/or local agro-ecological systems.
Eight PPV nucleotide sequences were reported, among which seven were new isolates
According to the above outcomes, PPV amplicons sequencing allowed to obtain 37 sequences, among which 26 belonged to PPV-M and 11 to PPV-Rec (Table 1). BLASTn analysis of PPV-M isolates allowed the identification of five new isolates (IT-AR 1, 2, 4, 5, IT-LU 1), as well as one isolate (IT-AR 3) identical (100.00% similarity) to isolate 04 (KF840166) previously reported in Slovakia on plum. Twenty-one sequences were identical to IT-AR 1, whereas only one sequence was associated to each of other isolates. IT-AR 1, 2, 4, and 5 showed high rates of similarity (99.75, 99.51, 99.50 and 99.27%, respectively) with PPV-Vegama (MW251494) previously found in Czech Republic on apricot cv. Vegama, whereas IT-LU 1 displayed 98.53% similarity with isolate Xi1/5 (MG458873) recovered in Bulgaria on plum. Importantly, IT-AR 1 was found in both peach and plum, whereas other PPV-M isolates were obtained only in peach. Differently, BLASTn analysis of PPV-Rec isolates allowed the identification of two isolates (IT-FI 1 and 2) occurring only in plum. Specifically, eight and three sequences were associated to IT-FI 1 and IT-FI 2, respectively, them showing a 99.11 and 98.93% similarity to isolate IT-ERMO-SH1907/08 (GU942476) previously detected in Italy on plum cv. Black Top. The identity levels of nucleotide and deduced amino acid sequences among PPV-M isolates were 97.71–99.75% and 95.59–100.00% (Table 2) and in PPV-Rec isolates were 99.82% and 100.00%, respectively. All identified PPV-M and PPV-Rec isolates were deposited in GenBank (www.ncbi.nlm.ni.gov).
Low genetic variability was reported in the identified PPV-M and PPV-Rec populations
The NIb/CP genetic variabilities (π) reported in PPV-M (0.010 ± 0.003) and in PPV-Rec (0.002 ± 0.002) populations were in accordance with low intra-strain diversity previously reported in PPV strains (García et al., 2014). Actually, π here reported are much lower than those reported by Kamenova and Borisova (2019) in Bulgaria (0.020 ± 0.004 and 0.018 ± 0.003, respectively), and even more than those emerging from the database here ad hoc developed to include sequences deposited in GenBank (Supplementary Tables 1, see Materials and Methods section for further details) referred to isolates from Mediterranean countries, namely Albania, Bosnia and Herzegovina, Croatia, France, Greece, Italy, Montenegro and Turkey (0.033 ± 0.004 and 0.026 ± 0.004, respectively). As virus isolates are commonly created through constant genetic drift and selection (LaTourette et al., 2022), the observed restricted π is likely due to efficacious monitoring and eradication activities (mandatory until 2021) to control PPV diffusion (Gougherty and Nutter, 2015).
PPV-Mb subgroup was firstly identified in Italy, and it was even prevalent than PPV-Ma
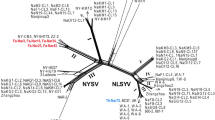
The positions of synonymous (8606 nt compared to M92280 isolate) and non-synonymous (8775 nt) nucleotide changes in CP region reported by Dallot et al. (2011) were observed in PPV-M isolates to differentiate between the subgroups Ma and Mb, which provenance is associated to Mediterranean and eastern European countries, respectively. The nucleotide changes showed IT-AR 1, 2, 3, 4, 5 belonging to subgroup Mb whereas IT-LU 1 to subgroup Ma. The phylogenetic analysis confirmed that PPV-M isolates were splitted in the two Ma and Mb clades supported by high bootstrap values (80 and 70%, respectively) among which the new isolates distributed in agreement with the previous analysis (Fig. 2). The present study, not only revealed both Ma and Mb subgroups, but unexpectedly Mb resulted prevalent as it was reported from five isolates (IT-AR 1, 2, 3, 4, 5) detected in Arezzo and Florence districts, whereas Ma was reported only in one isolate (IT-LU 1) detected in Lucca area. Differently, all the Italian isolates L9-1b (KJ994236), 9-335b (KJ994238), IT1 (EF626554), IT2 (EF626555), IT3 (EF626556) and CREA-DC_PPV6 (OL771187) previously reported in GenBank belong only to Ma. Therefore, to the best of our knowledge, this is the first record of Mb isolates in Italy.
Phylogenetic tree of plum pox virus Marcus (PPV-M) isolates from Italy reconstructed from partial NIb/CP genomic region. The trees were generated by Maximum Likelihood (ML), using the Jukes-Cantor model of evolution for nucleotide. The significance of each branch was evaluated by constructing 1,000 trees in bootstrap analysis. Bootstrap values are shown. The isolates sequenced in this study are in bold. The country of origin is shown in brackets after the name of the isolate. Subdivisions between strain and among PPV-M strain subgroups are reported on the right
PPV-Rec isolates were phylogenetically close to Italian and Turkish isolates previously detected
The phylogenetic analysis conducted using all the available PPV-M isolates containing NIb/CP genomic region retrieved in Genbank and included in the ad hoc developed database showed low bootstrap values among M strain and IT-AR 1, 2, 4, 5 resulted closed to the Bosnian isolate BOS150Pl (AJ749995) and to the German PPV-CGG-M7 (AY450597), found both in plum. IT-AR 3 was close to the Slovakian isolates 03 (KF840165) and 04 (KF840166), recovered in peach and plum, respectively, as well as to the North Macedonia isolates MK112 (MK562731) and MK175 (MK562732) found in peach and apricot, respectively. IT-LU 1 was close to the Turkish EdMrPl276 (MW415771) and North Macedonia MK41 (MK562730) isolates detected in plum and myrobalan, respectively (Fig. 3 and Supplementary Figure S1). Conversely, the analogous phylogenetic analysis for PPV-Rec strain showed high bootstrap values, with IT-FI 1 and 2 resulting close to the Italian isolates PPVBR (AJ812242) from Apulia and IT-ERMO-SH1907/08 (GU942476) from Emilia Romagna found in apricot and Japanese plum, respectively, as well as to the Turkish isolate Isparta (EF051630) identified in Japanese plum (Fig. 4 and Supplementary Figure S2). This result suggests a common origin for these isolates, and it may allow to hypothesize a propagation of PPV-Rec strains by movement of infected material along Italy and the Mediterranean basin (Sastry, 2012). Nevertheless, despite these high bootstrap values, no straight country or host-based clustering was observed among PPV-Rec population detected in the present study, as well as among PPV-M ones.
Portion of the phylogenetic tree of plum pox virus Marcus (PPV-M) reconstructed from partial NIb/CP genomic region. The trees were generated by Maximum Likelihood (ML), using the Jukes-Cantor model of evolution for nucleotide. The significance of each branch was evaluated by constructing 1,000 trees in bootstrap analysis. Bootstrap values are shown. The isolates sequenced in this study are in bold. The country of origin is shown in brackets after the name of the isolate. The complete PPV-M phylogenetic tree is available in Supplementary Figure S1
Portion of the phylogenetic tree of plum pox virus Recombinant (PPV-Rec) reconstructed from partial NIb/CP genomic region. The trees were generated by Maximum Likelihood (ML), using the Jukes-Cantor model of evolution for nucleotide. The significance of each branch was evaluated by constructing 1,000 trees in bootstrap analysis. Bootstrap values are shown. The isolates sequenced in this study are in bold. The country of origin is shown in brackets after the name of the isolate. The complete PPV-Rec phylogenetic tree is available in Supplementary Figure S2
Conclusions
Although in 2019 the European Union regraded PPV to RNQP due to its widespread endemic presence, this virus still represents one of the most devastating pathogen of stone fruits, with huge yield and economic losses worldwide. Thus, maintenance and improvement of diagnostic and molecular characterization activities is still crucial, especially considering that viruses and their vectors seem to be strongly favoured by climate change. This research provided a first broad molecular characterization of PPV strains in stone fruits of Tuscany. First, it confirmed PPV presence in this region, often occurring as co-infection with other viruses and viroids, and with PPV-M strain mostly reported in peach and PPV-Rec only detected in plum. Furthermore, starting from the identification of eight PPV nucleotide sequences (among which seven were new isolates), this study firstly identified the PPV-Mb subgroup in Italy, which was even prevalent than PPV-Ma. Finally, PPV-Rec isolates resulted phylogenetically close to Italian and Turkish isolates previously detected. Overall, the results here presented represent an important step to fill knowledge gaps about PPV in Tuscany, and we believe it may encourage other similar research to achieve more accurate data on PPV populations at both national and international levels.
References
Albert L, Willeit H, Steiner R (1974) Die Sharkakrankheit eineernste gefahr fùr die marillen bestatide im vinschgau. Obstauweinbau 11:314–315
Atanasoff D (1932) Plum pox. A new virus disease. Annals of the University of Sofia 11:49–69
Borisova A, Sotirov D (2021) The response of newly introduced plum cultivars to natural infection with Plum pox virus. Acta Horticulturae 1322:289–294. https://doi.org/10.17660/actahortic.2021.1322.40
Cambra M, Capote N, Myrta A, Llácer G (2006) Plum pox virus and the estimated costs associated with Sharka disease. EPPO Bulletin 36:202–204. https://doi.org/10.1111/j.13652338.2006.01027.x
Çelik A, Santosa AI, Gibbs AJ, Ertunç F (2022) Prunus necrotic ringspot virus in Turkey: an immigrant population. Archives of Virology 167:553–562. https://doi.org/10.1007/s00705-022-05374-1
Chirkov S, Sheveleva A, Ivanov P, Zakubanskiy A Analysis of genetic diversity of Russian sour cherry Plum pox virus isolates provides evidence of a new strain. Plant Disease 102:569–575., Akbilek A, Ilbağı Y (2018) H (2021) First report of Plum pox virus on Tilia spp. in Turkey. New Disease Reports 44. https://doi.org/10.1002/ndr2.12027
Dallot S, Glasa M, Jevremovic D, Kamenova I, Paunovic S, Labonne G (2011) Mediterranean and central-eastern european countries host viruses of two different clades of Plum pox virus strain M. Archives of Virology 156:539–542. https://doi.org/10.1007/s00705-011-0918-y
EPPO (2022) https://gd.eppo.int/taxon/PPV000/distribution
European Union (2019) Regulations-Commission implementing regulation (EU) 2019/2072 of 28 November 2019 establishing uniform conditions for the implementation of regulation (EU) 2016/2031 of the European Parliament and the Council, as regards protective measures against pests of plants, and repealing Commission Regulation (EC) no 690/2008 and amending Commission implementing regulation (EU) 2018/2019. Official Journal of the European Union 1:1–279
Faggioli F, Barba M (2006) Peach latent mosaic viroid: major findings of our studies over a period of fifteen years in Italy. Acta Horticulturae 781:529–534. https://doi.org/10.17660/actahortic.2008.781.77
Flores R, Delgado S, Rodio ME, Ambrós S, Hernández C, Serio FD (2006) Peach latent mosaic viroid: not so latent. Molecular Plant Pathology 7:209–221. https://doi.org/10.1111/j.1364-3703.2006.00332.x
García JA, Glasa M, Cambra M, Candresse T (2014) Plum pox virus and sharka: a model potyvirus and a major disease. Molecular Plant Pathology 15:226–241. https://doi.org/10.1111/mpp.12083
Ginanni M, Materazzi A, Mainardi M, Triolo E (1993) La “Vaiolatura” delle drupacee: indagini sulla presenza in Toscana. Informatore Fitopatologico 43(1):58–62
Glasa M, Prikhodko Y, Predajňa L, Nagyová A, Shneyder Y, Zhivaeva T, Šubr Z, Cambra M, Candresse T (2013) Characterization of Sour Cherry Isolates of Plum pox virus from the Volga Basin in Russia reveals a New Cherry strain of the Virus. Phytopathology 103:972–979. https://doi.org/10.1094/phyto-11-12-0285-r
Gospodaryk A, Moročko-Bičevska I, Pūpola N, Kāle A Occurrence of stone fruit viruses in plum orchards in Latvia. Proceedings of the Latvian Academy of Sciences. Section B., Natural (2013) Exact, and Applied Sciences 67:116–123. https://doi.org/10.2478/prolas-2013-0018
Gougherty AV, Nutter FW Jr (2015) Impact of eradication programs on the temporal and spatial dynamics of Plum pox virus on Prunus spp. in Pennsylvania and Ontario, Canada. Plant Disease 99:593–603. https://doi.org/10.1094/PDIS-03-14-0224-RE
Hadidi A, Barba M, Candresse T, Jelkmann W (2011) Virus and virus-like diseases of pome and stone fruits. Book 1:1–429. https://doi.org/10.1094/9780890545010
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposyum Series 41:95–98
ISTAT (2021) http://dati.istat.it/Index.aspx?QueryId=33705
James D, Thompson D (2006) Hosts and symptoms of Plum pox virus: ornamental and wild Prunus species. Bullettin OEPP/EPPO Bullettin 36:222–224. https://doi.org/10.1111/j.1365-2338.2006.00976.x
James D, Varga A, Sanderson D (2013) Genetic diversity of Plum pox virus: strains disease and related challenges for control. Canadian Journal of Plant Pathology 35(4) 431-441 10.1080/07060661.2013.828100
Kamenova I, Borisova A (2019) Update on distribution and genetic variability of Plum pox virus strains in Bulgaria. The Plant Pathology Journal 35:243–256. https://doi.org/10.5423/ppj.oa.09.2018.0189
Kim BT, Gibson PG, Scott SW (2010) Expression of the coat protein genes of PNRSV and PDV in the synergistic disease peach stunt-21st International Conference on Virus and other Graft Transmissible Diseases of Fruit Crops. Julius-Kühn-Archiv 427:114–117
Kinoti WM, Constable FE, Nancarrow N, Plummer KM, Rodoni B (2017) Analysis of intra-host genetic diversity of Prunus necrotic ringspot virus (PNRSV) using amplicon next generation sequencing. PLoS ONE 12:e0179284. https://doi.org/10.1371/journal.pone.0179284
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution 35:1547–1549. https://doi.org/10.1093/molbev/msy096
LaTourrette K, García-Ruiz H (2022) Determinants of virus variation, evolution, and host adaptation. Pathogens 11:1039. https://doi.org/10.3390/pathogens11091039
Li R, Mock R, Huang Q, Abad J, Hartung J, Kinard G (2008) A reliable and inexpensive method of nucleic acid extraction for the PCR-based detection of diverse plant pathogens. Journal of Virological Methods 154:48–55. https://doi.org/10.1016/j.jviromet.2008.09.008
Luigi M, Faggioli F (2011) Development of quantitative real-time RT-PCR for the detection and quantification of Peach latent mosaic viroid. European Journal of Plant Pathology 130:109–116. https://doi.org/10.1007/s10658-010-9738-2
Luigi M, Faggioli F (2013) Development of a quantitative real-time RT-PCR (qRT-PCR) for the detection of hop stunt viroid. European Journal of Plant Pathology 137:231–235. https://doi.org/10.1007/s10658-013-0243-2
Myrta A, Al Rwahnih M, Savino V (2005) Presence of a recombinant isolate of Plum pox virus in Apulia. Journal of Plant Pathology 87:127–130
Olmos A, Bertolini E, Gil M, Cambra M (2005) Real-time assay for quantitative detection of non-persistently transmitted Plum pox virus RNA targets in single aphids. Journal of Virological Methods 128:151–155. https://doi.org/10.1016/j.jviromet.2005.05.011
Osman F, Rowhani A (2008) Real-time RT-PCR (TaqMan®) assays for the detection of viruses associated with rugose wood complex of grapevine. Journal of Virological Methods 154:69–75. https://doi.org/10.1016/j.jviromet.2008.09.005
Osman F, Al Rwahnih M, Rowhani A (2017) Real-time RT-qPCR detection of Cherry rasp leaf virus, Cherry green ring mottle virus, Cherry necrotic rusty mottle virus, Cherry virus a and Apple chlorotic leaf spot virus in stone fruits. Journal of Plant Pathology 99:279–285. https://doi.org/10.4454/jpp.v99i1.3789
Pedrelli A, Panattoni A, Materazzi A (2021) Occurrence of Grapevine Pinot gris virus in Chianti vineyards. Agrochimica Special issue 65:81–87. https://doi.org/10.12871/00021857202201
Pigliónico D, Ojeda ME, Lucero V, Farrando R, Porcel L, Picca C, Marini D (2021) Spiraea sp. new natural host of Plum pox virus (sharka). European Journal of Plant Pathology 159:959–962. https://doi.org/10.1007/s10658-021-02206-x
Ragozzino E, Faggioli F, Barba M (2005) Detection and distribution of Citrus exocortis viroid and Hop stunt viroid in citrus orchards of Central Italy as revealed by one-tube one-step RT-PCR. International Organization of Citrus Virologists Conference Proceedings (1957–2010) 16. https://doi.org/10.5070/C59xn0r5td
Revers F, García JA (2015) Molecular biology of potyviruses. Advance in Virus Research 92:101 – 99. https://doi.org/10.1016/bs.aivir.2014.11.006
Rizza S, Conti F, Pasquini G, Tessitori M (2014) First report of Plum pox virus strain M isolates in Apricot in Sicily, Italy. Plant Disease 98:1591. https://doi.org/10.1094/PDIS-05-14-0458-PDN
Rizzo D, Materazzi A, Stefani L, Panattoni A, Pierro R, De Bellis L, Luvisi A (2017) The occurrence of viruses and viroids in ornamental citrus mother plants in Tuscany (Central Italy). Crop Protection 102:137–140. https://doi.org/10.1016/j.cropro.2017.08.026
Rubio M, Martínez-Gómez P, Marais A, Sánchez-Navarro J, Pallás V, Candresse T (2017) Recent advances and prospects in Prunus virology. Annals of Applied Biology 171:125–138. https://doi.org/10.1111/aab.12371
Salem N, Mansour A, Al-Musa A, Al-Nsour A (2003) Incidence of Prunus necrotic ringspot virus in Jordan. Phytopathologia Mediterranea 42:275–279. https://doi.org/10.14601/Phytopathol_Mediterr-1717
Schneider WL, Damsteegt VD, Gildow FE, Stone A L, Sherman DJ, Levy LE, Mavrodieva V, Richwine N, Welliver R, Luster DG (2011) Molecular ultrastructural and biological characterization of Pennsylvania isolates of Plum pox virus. Phytopathology 101(5) 627–636 10.1094/PHYTO-09-10-0256
Sebestyen D, Nemeth M, Hangyal R, Krizbai L, Ember I, Nyerges K, Kolber M, Kiss E, Bese G (2008) Ornamental Prunus species as new natural hosts of Plum pox virus and their importance in the spread of the virus in Hungary. Journal of Plant Pathology 90:S57–S61. https://www.jstor.org/stable/41998442
Serra P, Bertolini E, Martínez MC, Cambra M, Flores R (2017) Interference between variants of Peach latent mosaic viroid reveals novel features of its fitness landscape: implications for detection. Scientific Reports 7. https://doi.org/10.1038/srep42825
Sihelská N, Glasa M, Šubr ZW (2017) Host preference of the major strains of Plum pox virus - opinions based on regional and world-wide sequence data. Journal of Integrative Agriculture 16:510–515. https://doi.org/10.1016/S2095-3119(16)61356-4
Sochor J, Babula P, Adam V, Krska B, Kizek R (2012) Sharka: the past, the present and the future. Viruses 4:2853–2901. https://doi.org/10.3390/v4112853
Subr Z, Pittnerová S, Glasa M (2004) A simplified RT-PCR-based detection of recombinant Plum pox virus isolates. Acta Virologica 48:173–176
Sastry KS (2012) Seed-borne plant virus diseases Plant Virus Transmission through vegetative propagules (asexual reproduction). Seed-Borne Plant Virus Diseases 1: 285–305.
Yardimci BCN, Culal-Klllc H (2011) Detection of viruses infecting stone fruits in western Mediterranean region of Turkey. The Plant Pathology Journal 27:44–52. https://doi.org/10.5423/ppj.2011.27.1.044
Zagrai LA, Zagrai I, Guzu GM, Roșu-Mareș SD, Moldovan C (2022) Assessment of the virus infections occurrence in new established plum and sweet cherry orchards in Transylvania, Romania. Notulae Botanicae Horti Agrobotanici Cluj-Napoca 50:12734. https://doi.org/10.15835/nbha50212734
Acknowledgements
We gratefully acknowledge the Regional Phytosanitary Service of Tuscany for their precious support in field sampling.
Funding
Open access funding provided by Università di Pisa within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pedrelli, A., Panattoni, A. & Cotrozzi, L. First molecular characterization of plum pox virus strains in stone fruits of Tuscany (Central Italy). J Plant Pathol 105, 1045–1053 (2023). https://doi.org/10.1007/s42161-023-01430-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42161-023-01430-0