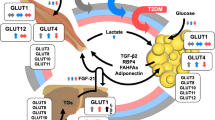
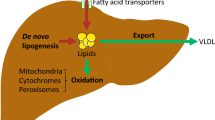
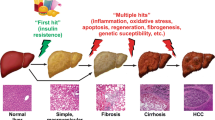
Abstract
Obesity and its associated metabolic derangements have become a major global health challenge. Ectopic fat accumulation disrupts metabolic homeostasis leading to metabolic syndrome (MetS), type 2 diabetes (T2D), and cardiovascular diseases. T2D is strongly associated with chronic low-grade inflammation in the adipose tissue, liver, and arguably in the skeletal muscle. Secretory proteins elaborated by these organs, i.e., adipokines, hepatokines, and myokines, are collectively grouped as organokines, which interact with each other to produce complex effects in insulin target tissues through endocrine, autocrine, and paracrine pathways. Since organokines have both proinflammatory and anti-inflammatory effects, the optimum balance between them is critical for metabolic homeostasis. The goal of this review is to focus on the functions of some of these organokines that have been identified in contemporary research as major regulators of inflammation, leading to the onset and progression of metabolic diseases.




Similar content being viewed by others
Abbreviations
- ACC:
-
Acetyl-CoA carboxylase
- AdipoR:
-
Adiponectin receptor
- ADSF:
-
Adipocyte-specific secretory factor
- AHSG:
-
α-2-Heremans-Schmid glycoprotein
- α-MSH:
-
α-Melanocyte-stimulating hormone
- AMPK:
-
AMP-activated protein kinase
- aP2:
-
Adipocyte protein 2
- ApoER2:
-
Apolipoprotein E receptor 2
- APPL1:
-
Adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1
- ATM:
-
Adipose tissue macrophages
- BMI:
-
Body mass index
- CaMKKβ:
-
Calcium/calmodulin-dependent protein kinase kinase-β
- CAP1:
-
Cyclase-associated protein 1
- CCL2:
-
Chemokine C–C motif ligand 2
- CCR2:
-
C–C chemokine receptor type 2
- CD36:
-
Cluster of differentiation 36
- ChemR23:
-
Chemerin receptor 23
- CMKLR1:
-
Chemokine-like receptor 1
- CPT1:
-
Carnitine palmitoyltransferase I
- CREB:
-
cAMP-response element-binding protein
- CRTC2:
-
CREB-regulated transcription coactivator 2
- DAG:
-
Diacylglycerol
- DPP-IV:
-
Dipeptidyl peptidase 4
- ERK:
-
Extracellular signal-regulated kinase
- FAT:
-
Fatty acid translocase
- FetA:
-
Fetuin-A
- FGF21:
-
Fibroblast growth factor-21
- FIZZ:
-
Found in inflammatory zone
- FOXO:
-
Forkhead box protein O
- G6Pase:
-
Glucose-6-phosphatase
- GLUT:
-
Glucose transporter
- GSK3B:
-
Glycogen synthase kinase-3 β
- GYS:
-
Glycogen synthase
- HFREP1:
-
Hepatocyte-derived fibrinogen-related protein 1
- HIF-1α:
-
Hypoxia-inducible factor-1α
- HNF-4α:
-
Hepatocyte nuclear factor-4α
- HO-1:
-
Heme oxygenase-1
- HOMA-IR:
-
Homeostasis model assessment-estimated insulin resistance
- IFNγ:
-
Interferon γ
- IL:
-
Interleukin
- IL-6R:
-
Interleukin-6 receptor
- iNOS:
-
Inducible nitric oxide synthase
- IRS:
-
Insulin receptor substrate
- IRβ:
-
Insulin receptor β
- ISL-1:
-
Insulin gene enhancer protein-1
- JAK:
-
Janus kinase
- JNK:
-
c-Jun N-terminal kinase
- Keap1:
-
Kelch-like ECH-associated protein 1
- LEPR:
-
Leptin receptor
- LKB1:
-
Liver kinase B1
- MAF:
-
Monocyte activating factor
- MAPK:
-
Mitogen-activated protein kinase
- MCF:
-
Monocyte chemotactic factor
- MCP-1:
-
Monocyte chemoattractant protein-1
- MD2:
-
Myeloid differentiation factor 2
- MEK:
-
Mitogen-activated protein kinase kinase
- MetS:
-
Metabolic syndrome
- mTORC1:
-
Mammalian target of rapamycin complex 1
- MyD88:
-
Myeloid differentiation primary response 88
- NF-κB:
-
Nuclear factor-κB
- NOX:
-
NADPH Oxidase
- Nrf2:
-
Nuclear factor erythroid 2-related factor 2
- PAI-1:
-
Plasminogen activator inhibitor-1
- PEPCK:
-
Phosphoenolpyruvate carboxykinase
- PGC-1α:
-
Peroxisome proliferator-activated receptor-gamma coactivator-1α
- PI3K:
-
Phosphoinositide 3-kinase
- PKA:
-
Protein kinase A
- PKB:
-
Protein kinase B
- PKC:
-
Protein kinase C
- POMC:
-
Pro-opiomelanocortin
- PP2A:
-
Protein phosphatase 2A
- PPARγ:
-
Peroxisome proliferator-activated receptor γ
- PTEN:
-
Phosphatase and tensin homolog
- PTP1B:
-
Protein tyrosine phosphatases-1B
- Ras:
-
Rat sarcoma virus
- RBP4:
-
Retinol-binding protein 4
- RELMs:
-
Resistin-like molecules
- ROS:
-
Reactive oxygen species
- SeP:
-
Selenoprotein P
- Ser:
-
Serine
- SFA:
-
Saturated fatty acids
- SHBG:
-
Sex hormone-binding globulin
- sIL-6R:
-
Soluble interleukin-6 receptor
- SIRT1:
-
Sirtuin1
- SOCS3:
-
Suppressor of cytokine signaling 3
- SREBP1c:
-
Sterol regulatory element-binding protein 1c
- STAT:
-
Signal transducers and activators of transcription
- STRA6:
-
Stimulated by retinoic acid 6
- T2D:
-
Type 2 diabetes
- TGFβ:
-
Transforming growth factor-β
- TLR:
-
Toll-like receptor
- TNFR:
-
Tumor necrosis factor receptor
- TNFα:
-
Tumor necrosis factor α
- tPA:
-
Tissue plasminogen activator
- TRIF:
-
TIR-domain-containing adapter- inducing interferon-β
- TSC2:
-
Tuberous sclerosis complex 2
- Tyr:
-
Tyrosine
- uPA:
-
Urokinase plasminogen activator
- VCAM-1:
-
Vascular cell adhesion molecule 1
- Wnt3a:
-
Wnt family member 3a
- XCP1:
-
Cysteine-rich protein 1
References
IDF Diabetes Atlas. 10th edition, International Diabetes Federation, 2022. https://diabetesatlas.org/. Accessed 21 Dec 2022
Oda E (2018) Historical perspectives of the metabolic syndrome. Clin Dermatol 36:3–8. https://doi.org/10.1016/j.clindermatol.2017.09.002
Shin JA, Lee JH, Lim SY, Ha HS, Kwon HS, Park YM, Lee WC, Kang MI, Yim HW, Yoon KH, Son HY (2013) Metabolic syndrome as a predictor of type 2 diabetes, and its clinical interpretations and usefulness. J Diabetes Investig 4:334–343. https://doi.org/10.1111/jdi.12075
Krishnamoorthy Y, Rajaa S, Murali S, Rehman T, Sahoo J, Kar SS (2020) Prevalence of metabolic syndrome among adult population in India: a systematic review and meta-analysis. PLoS ONE 15:e0240971. https://doi.org/10.1371/journal.pone.0240971
Krupp K, Adsul P, Wilcox ML, Srinivas V, Frank E, Srinivas A, Madhivanan P (2020) Prevalence and correlates of metabolic syndrome among rural women in Mysore, India. Indian Heart J 72:582–588. https://doi.org/10.1016/j.ihj.2020.09.015
Must A, Spadano J, Coakley EH, Field AE, Colditz G, Dietz WH (1999) The disease burden associated with overweight and obesity. JAMA 282:1523–1529. https://doi.org/10.1001/jama.282.16.1523
Reddy P, Lent-Schochet D, Ramakrishnan N, McLaughlin M, Jialal I (2019) Metabolic syndrome is an inflammatory disorder: a conspiracy between adipose tissue and phagocytes. Clin Chim Acta 496:35–44. https://doi.org/10.1016/j.cca.2019.06.019
Hotamisligil GS, Shargill NS, Spiegelman BM (1993) Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science 259:87–91. https://doi.org/10.1126/science.7678183
Hotamisligil GS, Arner P, Caro JF, Atkinson RL, Spiegelman BM (1995) Increased adipose tissue expression of tumor necrosis factor-alpha in human obesity and insulin resistance. J Clin Investig 95:2409–2415. https://doi.org/10.1172/JCI117936
Lee YS, Olefsky J (2021) Chronic tissue inflammation and metabolic disease. Genes Dev 35:307–328. https://doi.org/10.1101/gad.346312.120
Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW Jr (2003) Obesity is associated with macrophage accumulation in adipose tissue. J Clin Investig 112:1796–1808. https://doi.org/10.1172/JCI19246
Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, Chen H (2003) Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Investig 112:1821–1830. https://doi.org/10.1172/JCI19451
Lumeng CN, Bodzin JL, Saltiel AR (2007) Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Investig 117:175–184. https://doi.org/10.1172/JCI29881
Lumeng CN, Deyoung SM, Bodzin JL, Saltiel AR (2007) Increased inflammatory properties of adipose tissue macrophages recruited during diet-induced obesity. Diabetes 56:16–23. https://doi.org/10.2337/db06-1076
Lee YH, Petkova AP, Granneman JG (2013) Identification of an adipogenic niche for adipose tissue remodeling and restoration. Cell Metab 18:355–367. https://doi.org/10.1016/j.cmet.2013.08.003
Olefsky JM, Glass CK (2010) Macrophages, inflammation, and insulin resistance. Annu Rev Physiol 72:219–246. https://doi.org/10.1146/annurev-physiol-021909-135846
Coats BR, Schoenfelt KQ, Barbosa-Lorenzi VC, Peris E, Cui C, Hoffman A, Zhou G, Fernandez S, Zhai L, Hall BA, Haka AS, Shah AM, Reardon CA, Brady MJ, Rhodes CJ, Maxfield FR, Becker L (2017) Metabolically activated adipose tissue macrophages perform detrimental and beneficial functions during diet-induced obesity. Cell Rep 20:3149–3161. https://doi.org/10.1016/j.celrep.2017.08.096
Hill DA, Lim HW, Kim YH, Ho WY, Foong YH, Nelson VL, Nguyen HCB, Chegireddy K, Kim J, Habertheuer A, Vallabhajosyula P, Kambayashi T, Won KJ, Lazar MA (2018) Distinct macrophage populations direct inflammatory versus physiological changes in adipose tissue. Proc Natl Acad Sci USA 115:5096–5105. https://doi.org/10.1073/pnas.1802611115
Jaitin DA, Adlung L, Thaiss CA, Weiner A, Li B, Descamps H, Lundgren P, Bleriot C, Liu Z, Deczkowska A, Keren-Shaul H, David E, Zmora N, Eldar SM, Lubezky N, Shibolet O, Hill DA, Lazar MA, Colonna M, Ginhoux F, Shapiro H, Elinav E, Amit I (2019) Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell 178:686–698. https://doi.org/10.1016/j.cell.2019.05.054
Kratz M, Coats BR, Hisert KB, Hagman D, Mutskov V, Peris E, Schoenfelt KQ, Kuzma JN, Larson I, Billing PS, Landerholm RW, Crouthamel M, Gozal D, Hwang S, Singh PK, Becker L (2014) Metabolic dysfunction drives a mechanistically distinct proinflammatory phenotype in adipose tissue macrophages. Cell Metab 20:614–625. https://doi.org/10.1016/j.cmet.2014.08.010
Xu X, Grijalva A, Skowronski A, van Eijk M, Serlie MJ, Ferrante AW Jr (2013) Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab 18:816–830. https://doi.org/10.1016/j.cmet.2013.11.001
Cinti S, Mitchell G, Barbatelli G, Murano I, Ceresi E, Faloia E, Wang S, Fortier M, Greenberg AS, Obin MS (2005) Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J Lipid Res 46:2347–2355. https://doi.org/10.1194/jlr.M500294-JLR200
Pal D, Dasgupta S, Kundu R, Maitra S, Das G, Mukhopadhyay S, Ray S, Majumdar SS, Bhattacharya S (2012) Fetuin-A acts as an endogenous ligand of TLR4 to promote lipid-induced insulin resistance. Nat Med 18:1279–1285. https://doi.org/10.1038/nm.2851
Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM (1994) Positional cloning of the mouse obese gene and its human homologue. Nature 372:425–432. https://doi.org/10.1038/372425a0
Ingalls AM, Dickie MM, Snell GD (1950) Obese, a new mutation in the house mouse. J Hered 41:317–318. https://doi.org/10.1093/oxfordjournals.jhered.a106073
Cowley MA, Smart JL, Rubinstein M, Cerdán MG, Diano S, Horvath TL, Cone RD, Low MJ (2001) Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature 411:480–484. https://doi.org/10.1038/35078085
Maffei M, Halaas J, Ravussin E, Pratley RE, Lee GH, Zhang Y, Fei H, Kim S, Lallone R, Ranganathan S, Kern PA, Friedman JM (1995) Leptin levels in human and rodent: measurement of plasma leptin and ob RNA in obese and weight-reduced subjects. Nat Med 1:1155–1161. https://doi.org/10.1038/nm1195-1155
Tartaglia LA, Dembski M, Weng X, Deng N, Culpepper J, Devos R, Richards GJ, Campfield LA, Clark FT, Deeds J, Muir C, Sanker S, Moriarty A, Moore KJ, Smutko JS, Mays GG, Wool EA, Monroe CA, Tepper RI (1995) Identification and expression cloning of a leptin receptor, OB-R. Cell 83:1263–1271. https://doi.org/10.1016/0092-8674(95)90151-5
Facey A, Dilworth L, Irving R (2017) A review of the leptin hormone and the association with obesity and diabetes mellitus. J Diabetes Metab. https://doi.org/10.4172/2155-6156.1000727
Pérez-Pérez A, Sánchez-Jiménez F, Vilariño-García T, Sánchez-Margalet V (2020) Role of leptin in inflammation and vice versa. Int J Mol Sci 21:5887. https://doi.org/10.3390/ijms21165887
Deb A, Deshmukh B, Ramteke P, Bhati FK, Bhat MK (2021) Resistin: a journey from metabolism to cancer. Transl Oncol 14:101178. https://doi.org/10.1016/j.tranon.2021.101178
Barnes KM, Miner JL (2009) Role of resistin in insulin sensitivity in rodents and humans. Curr Protein Pept 10:96–107. https://doi.org/10.2174/138920309787315239
Luo Z, Zhang Y, Li F, He J, Ding H, Yan L, Cheng H (2009) Resistin induces insulin resistance by both AMPK-dependent and AMPK-independent mechanisms in HepG2 cells. Endocrine 36:60–69. https://doi.org/10.1007/s12020-009-9198-7
Taouis M, Benomar Y (2021) Is resistin the master link between inflammation and inflammation-related chronic diseases? Mol Cell Endocrinol 533:111341. https://doi.org/10.1016/j.mce.2021.111341
Pandey GK, Vadivel S, Raghavan S, Mohan V, Balasubramanyam M, Gokulakrishnan K (2019) High molecular weight adiponectin reduces glucolipotoxicity-induced inflammation and improves lipid metabolism and insulin sensitivity via APPL1-AMPK-GLUT4 regulation in 3T3-L1 adipocytes. Atherosclerosis 288:67–75. https://doi.org/10.1016/j.atherosclerosis.2019.07.011
Achari AE, Jain SK (2017) Adiponectin, a therapeutic target for obesity, diabetes, and endothelial dysfunction. Int J Mol Sci 18:1321. https://doi.org/10.3390/ijms18061321
Ryu J, Galan AK, Xin X, Dong F, Abdul-Ghani MA, Zhou L, Wang C, Li C, Holmes BM, Sloane LB, Austad SN, Guo S, Musi N, DeFronzo RA, Deng C, White MF, Liu F, Dong LQ (2014) APPL1 potentiates insulin sensitivity by facilitating the binding of IRS1/2 to the insulin receptor. Cell Rep 7:1227–1238. https://doi.org/10.1016/j.celrep.2014.04.006
Iwabu M, Yamauchi T, Okada-Iwabu M, Sato K, Nakagawa T, Funata M, Yamaguchi M, Namiki S, Nakayama R, Tabata M, Ogata H, Kubota N, Takamoto I, Hayashi YK, Yamauchi N, Waki H, Fukayama M, Nishino I, Tokuyama K, Ueki K, Oike Y, Ishii S, Hirose K, Shimizu T, Touhara K, Kadowaki T (2010) Adiponectin and AdipoR1 regulate PGC-1alpha and mitochondria by Ca(2+) and AMPK/SIRT1. Nature 464:1313–1319. https://doi.org/10.1038/nature08991
Awazawa M, Ueki K, Inabe K, Yamauchi T, Kaneko K, Okazaki Y, Bardeesy N, Ohnishi S, Nagai R, Kadowaki T (2009) Adiponectin suppresses hepatic SREBP1c expression in an AdipoR1/LKB1/AMPK dependent pathway. Biochem Biophys Res Commun 382:51–56. https://doi.org/10.1016/j.bbrc.2009.02.131
Ceddia RB, Somwar R, Maida A, Fang X, Bikopoulos G, Sweeney G (2005) Globular adiponectin increases GLUT4 translocation and glucose uptake but reduces glycogen synthesis in rat skeletal muscle cells. Diabetologia 48:132–139. https://doi.org/10.1007/s00125-004-1609-y
Ryu J, Hadley JT, Li Z, Dong F, Xu H, Xin X, Zhang Y, Chen C, Li S, Guo X, Zhao JL, Leach RJ, Abdul-Ghani MA, DeFronzo RA, Kamat A, Liu F, Dong LQ (2021) Adiponectin alleviates diet-induced inflammation in the liver by suppressing MCP-1 expression and macrophage infiltration. Diabetes 70:1303–1316. https://doi.org/10.2337/db20-1073
Ma L, Xu Y, Zhang Y, Ji T, Li Y (2020) Lower levels of circulating adiponectin in elderly patients with metabolic inflammatory syndrome: a cross-sectional study. Diabet Metab Synd Ob 13:591–596. https://doi.org/10.2147/DMSO.S242397
Senthilkumar GP, Anithalekshmi MS, Yasir M, Parameswaran S, Packirisamy RM, Bobby Z (2018) Role of omentin 1 and IL-6 in type 2 diabetes mellitus patients with diabetic nephropathy. Diabetes Metab Syndr 12:23–26. https://doi.org/10.1016/j.dsx.2017.08.005
Schäffler A, Neumeier M, Herfarth H, Fürst A, Schölmerich J, Büchler C (2005) Genomic structure of human omentin, a new adipocytokine expressed in omental adipose tissue. Biochim Biophys Acta 1732:96–102. https://doi.org/10.1016/j.bbaexp.2005.11.005
Yang RZ, Lee MJ, Hu H, Pray J, Wu HB, Hansen BC, Shuldiner AR, Fried SK, McLenithan JC, Gong DW (2006) Identification of omentin as a novel depot-specific adipokine in human adipose tissue: possible role in modulating insulin action. Am J Physiol Endocrinol Metab 290:E1253–E1261. https://doi.org/10.1152/ajpendo.00572.2004
de Souza Batista CM, Yang RZ, Lee MJ, Glynn NM, Yu DZ, Pray J, Ndubuizu K, Patil S, Schwartz A, Kligman M, Fried SK, Gong DW, Shuldiner AR, Pollin TI, McLenithan JC (2007) Omentin plasma levels and gene expression are decreased in obesity. Diabetes 56:1655–1661. https://doi.org/10.2337/db06-1506
Kocijancic M, Vujicic B, Racki S, Cubranic Z, Zaputovic L, Dvornik S (2015) Serum omentin-1 levels as a possible risk factor of mortality in patients with diabetes on haemodialysis. Diabetes Res Clin Pract 110:44–50. https://doi.org/10.1016/j.diabres.2015.06.008
Onur I, Oz F, Yildiz S, Kuplay H, Yucel C, Sigirci S, Elitok A, Pilten S, Kasali K, Yasar Cizgici A, Erentug V, Dinckal HM (2014) A decreased serum omentin-1 level may be an independent risk factor for peripheral arterial disease. Int Angiol 33:455–460
Kazama K, Usui T, Okada M, Hara Y, Yamawaki H (2012) Omentin plays an anti-inflammatory role through inhibition of TNF-α-induced superoxide production in vascular smooth muscle cells. Eur J Pharmacol 686:116–123. https://doi.org/10.1016/j.ejphar.2012.04.033
Pan HY, Guo L, Li Q (2010) Changes of serum omentin-1 levels in normal subjects and in patients with impaired glucose regulation and with newly diagnosed and untreated type 2 diabetes. Diabetes Res Clin Pract 88:29–33. https://doi.org/10.1016/j.diabres.2010.01.013
Zhang Q, Zhu L, Zheng M, Fan C, Li Y, Zhang D, He Y, Yang H (2014) Changes of serum omentin-1 levels in normal subjects, type 2 diabetes and type 2 diabetes with overweight and obesity in Chinese adults. Ann Endocrinol (Paris) 75:171–175. https://doi.org/10.1016/j.ando.2014.04.013
Tan BK, Pua S, Syed F, Lewandowski KC, O’Hare JP, Randeva HS (2008) Decreased plasma omentin-1 levels in Type 1 diabetes mellitus. Diabet Med 25:1254–1255. https://doi.org/10.1111/j.1464-5491.2008.02568.x
Wittamer V, Franssen JD, Vulcano M, Mirjolet JF, Le Poul E, Migeotte I, Brézillon S, Tyldesley R, Blanpain C, Detheux M, Mantovani A, Sozzani S, Vassart G, Parmentier M, Communi D (2003) Specific recruitment of antigen-presenting cells by chemerin, a novel processed ligand from human inflammatory fluids. J Exp Med 198:977–985. https://doi.org/10.1084/jem.20030382
Wittamer V, Bondue B, Guillabert A, Vassart G, Parmentier M, Communi D (2005) Neutrophil- mediated maturation of chemerin: a link between innate and adaptive immunity. J Immunol 175:487–493. https://doi.org/10.4049/jimmunol.175.1.487
Goralski KB, McCarthy TC, Hanniman EA, Zabel BA, Butcher EC, Parlee SD, Muruganandan S, Sinal CJ (2007) Chemerin, a novel adipokine that regulates adipogenesis and adipocyte metabolism. J Biol Chem 282:28175–28188. https://doi.org/10.1074/jbc.M700793200
Takahashi M, Okimura Y, Iguchi G, Nishizawa H, Yamamoto M, Suda K, Kitazawa R, Fujimoto W, Takahashi K, Zolotaryov FN, Hong KS, Kiyonari H, Abe T, Kaji H, Kitazawa S, Kasuga M, Chihara K, Takahashi Y (2011) Chemerin regulates β-cell function in mice. Sci Rep 1:123. https://doi.org/10.1038/srep00123
Brunetti L, Orlando G, Ferrante C, Recinella L, Leone S, Chiavaroli A, Di Nisio C, Shohreh R, Manippa F, Ricciuti A, Vacca M (2014) Peripheral chemerin administration modulates hypothalamic control of feeding. Peptides 51:115–121. https://doi.org/10.1016/j.peptides.2013.11.007
Sell H, Laurencikiene J, Taube A, Eckardt K, Cramer A, Horrighs A, Arner P, Eckel J (2009) Chemerin is a novel adipocyte-derived factor inducing insulin resistance in primary human skeletal muscle cells. Diabetes 58:2731–2740. https://doi.org/10.2337/db09-0277
Roman AA, Parlee SD, Sinal CJ (2012) Chemerin: a potential endocrine link between obesity and type 2 diabetes. Endocrine 42:243–251. https://doi.org/10.1007/s12020-012-9698-8
Adeghate E (2008) Visfatin: structure, function and relation to diabetes mellitus and other dysfunctions. Curr Med Chem 15:1851–1862. https://doi.org/10.2174/092986708785133004
Fukuhara A, Matsuda M, Nishizawa M, Segawa K, Tanaka M, Kishimoto K, Matsuki Y, Murakami M, Ichisaka T, Murakami H, Watanabe E, Takagi T, Akiyoshi M, Ohtsubo T, Kihara S, Yamashita S, Makishima M, Funahashi T, Yamanaka S, Hiramatsu R, Matsuzawa Y, Shimomura I (2005) Visfatin: a protein secreted by visceral fat that mimics the effects of insulin. Science 307:426–430 [Retraction in Science, 318:565, 2007]. https://doi.org/10.1126/science.1097243
Heo YJ, Choi SE, Jeon JY, Han SJ, Kim DJ, Kang Y, Lee KW, Kim HJ (2019) Visfatin induces inflammation and insulin resistance via the NF-κB and STAT3 signaling pathways in hepatocytes. J Diabetes Res 2019:4021623. https://doi.org/10.1155/2019/4021623
Hirano T, Taga T, Nakano N, Yasukawa K, Kashiwamura S, Shimizu K, Nakajima K, Pyun KH, Kishimoto T (1985) Purification to homogeneity and characterization of human B-cell differentiation factor (BCDF or BSFp-2). Proc Natl Acad Sci USA 82:5490–5494. https://doi.org/10.1073/pnas.82.16.5490
Poupart P, Vandenabeele P, Cayphas S, Van Snick J, Haegeman G, Kruys V, Fiers W, Content J (1987) B cell growth modulating and differentiating activity of recombinant human 26-kd protein (BSF-2, HuIFN-beta 2, HPGF). EMBO J 6:1219–1224. https://doi.org/10.1002/j.1460-2075.1987.tb02357.x
Mohamed-Ali V, Goodrick S, Rawesh A, Katz DR, Miles JM, Yudkin JS, Klein S, Coppack SW (1997) Subcutaneous adipose tissue releases interleukin-6, but not tumor necrosis factor-alpha, in vivo. J Clin Endocrinol Metab 82:4196–4200. https://doi.org/10.1210/jcem.82.12.4450
Akbari M, Hassan-Zadeh V (2018) IL-6 signalling pathways and the development of type 2 diabetes. Inflammopharmacology 26:685–698. https://doi.org/10.1007/s10787-018-0458-0
Kraakman MJ, Kammoun HL, Allen TL, Deswaerte V, Henstridge DC, Estevez E, Matthews VB, Neill B, White DA, Murphy AJ, Peijs L, Yang C, Risis S, Bruce CR, Du XJ, Bobik A, Lee-Young RS, Kingwell BA, Vasanthakumar A, Shi W, Kallies A, Lancaster GI, Rose-John S, Febbraio MA (2015) Blocking IL-6 trans-signaling prevents high-fat diet-induced adipose tissue macrophage recruitment but does not improve insulin resistance. Cell Metab 21:403–416. https://doi.org/10.1016/j.cmet.2015.02.006
Rehman K, Akash MSH, Liaqat A, Kamal S, Qadir MI, Rasul A (2017) Role of interleukin-6 in development of insulin resistance and type 2 diabetes mellitus. Crit Rev Eukaryot 27:229–236. https://doi.org/10.1615/CritRevEukaryotGeneExpr.2017019712
van Meijer M, Pannekoek H (1995) Structure of plasminogen activator inhibitor 1 (PAI-1) and its function in fibrinolysis: an update. Fibrinolysis 9:263–276. https://doi.org/10.1016/S0268-9499(95)80015-8
Loskutoff DJ, van Mourik JA, Erickson LA, Lawrence D (1983) Detection of an unusually stable fibrinolytic inhibitor produced by bovine endothelial cells. Proc Natl Acad Sci USA 80:2956–2960. https://doi.org/10.1073/pnas.80.10.2956
Simpson AJ, Booth NA, Moore NR, Bennett B (1991) Distribution of plasminogen activator inhibitor (PAI-1) in tissues. J Clin Pathol 44:139–143. https://doi.org/10.1136/jcp.44.2.139
Alessi MC, Peiretti F, Morange P, Henry M, Nalbone G, Juhan-Vague I (1997) Production of plasminogen activator inhibitor 1 by human adipose tissue: possible link between visceral fat accumulation and vascular disease. Diabetes 46:860–867. https://doi.org/10.2337/diab.46.5.860
Correia ML, Haynes WG (2006) A role for plasminogen activator inhibitor-1 in obesity: from pie to PAI? Arterioscler Thromb Vasc Biol 26:2183–2185. https://doi.org/10.1161/01.ATV.0000244018.24120.70
Al-Hamodi Z, Ismail IS, Saif-Ali R, Ahmed KA, Muniandy S (2011) Association of plasminogen activator inhibitor-1 and tissue plasminogen activator with type 2 diabetes and metabolic syndrome in Malaysian subjects. Cardiovasc Diabetol 10:23. https://doi.org/10.1186/1475-2840-10-23
Jung RG, Motazedian P, Ramirez FD, Simard T, Di Santo P, Visintini S, Faraz MA, Labinaz A, Jung Y, Hibbert B (2018) Association between plasminogen activator inhibitor-1 and cardiovascular events: a systematic review and meta-analysis. Thromb J 16:12. https://doi.org/10.1186/s12959-018-0166-4
Samad F, Uysal KT, Wiesbrock SM, Pandey M, Hotamisligil GS, Loskutoff DJ (1999) Tumor necrosis factor alpha is a key component in the obesity-linked elevation of plasminogen activator inhibitor 1. Proc Natl Acad Sci USA 96:6902–6907. https://doi.org/10.1073/pnas.96.12.6902
Tang S, Liu W, Pan X, Liu L, Yang Y, Wang D, Xu P, Huang M, Chen Z (2020) Specific inhibition of plasminogen activator inhibitor 1 reduces blood glucose level by lowering TNF-a. Life Sci 246:117404. https://doi.org/10.1016/j.lfs.2020.117404
Nawaz SS, Siddiqui K (2022) Plasminogen activator inhibitor-1 mediate downregulation of adiponectin in type 2 diabetes patients with metabolic syndrome. Cytokine X 4:100064. https://doi.org/10.1016/j.cytox.2022.100064
Levine JA, Olivares S, Miyata T, Vaughan DE, Henkel AS (2021) Inhibition of PAI-1 promotes lipolysis and enhances weight loss in obese mice. Obesity (Silver Spring) 29:713–720. https://doi.org/10.1002/oby.23112
Kanai M, Raz A, Goodman DS (1968) Retinol-binding protein: the transport protein for vitamin A in human plasma. J Clin Investig 47:2025–2044. https://doi.org/10.1172/JCI105889
Thompson SJ, Sargsyan A, Lee SA, Yuen JJ, Cai J, Smalling R, Ghyselinck N, Mark M, Blaner WS, Graham TE (2017) Hepatocytes are the principal source of circulating RBP4 in mice. Diabetes 66:58–63. https://doi.org/10.2337/db16-0286
Yao-Borengasser A, Varma V, Bodles AM, Rasouli N, Phanavanh B, Lee MJ, Starks T, Kern LM, Spencer HJ 3rd, Rashidi AA, McGehee RE Jr, Fried SK, Kern PA (2007) Retinol binding protein 4 expression in humans: relationship to insulin resistance, inflammation, and response to pioglitazone. J Clin Endocrinol Metab 92:2590–2597. https://doi.org/10.1210/jc.2006-0816
Norseen J, Hosooka T, Hammarstedt A, Yore MM, Kant S, Aryal P, Kiernan UA, Phillips DA, Maruyama H, Kraus BJ, Usheva A, Davis RJ, Smith U, Kahn BB (2012) Retinol-binding protein 4 inhibits insulin signaling in adipocytes by inducing proinflammatory cytokines in macrophages through a c-Jun N-terminal kinase- and toll-like receptor 4-dependent and retinol-independent mechanism. Mol Cell Biol 32:2010–2019. https://doi.org/10.1128/MCB.06193-11
Kilicarslan M, de Weijer BA, SimonytéSjödin K, Aryal P, Ter Horst KW, Cakir H, Romijn JA, Ackermans MT, Janssen IM, Berends FJ, van de Laar AW, Houdijk AP, Kahn BB, Serlie MJ (2020) RBP4 increases lipolysis in human adipocytes and is associated with increased lipolysis and hepatic insulin resistance in obese women. FASEB J 34:6099–6110. https://doi.org/10.1096/fj.201901979RR
Huang R, Bai X, Li X, Wang X, Zhao L (2021) Retinol-binding protein 4 activates STRA6, provoking pancreatic β-cell dysfunction in type 2 diabetes. Diabetes 70:449–463. https://doi.org/10.2337/db19-1241
Matsushima K, Larsen CG, DuBois GC, Oppenheim JJ (1989) Purification and characterization of a novel monocyte chemotactic and activating factor produced by a human myelomonocytic cell line. J Exp Med 169:1485–1490. https://doi.org/10.1084/jem.169.4.1485
Yoshimura T, Robinson EA, Tanaka S, Appella E, Kuratsu J, Leonard EJ (1989) Purification and amino acid analysis of two human glioma-derived monocyte chemoattractants. J Exp Med 169:1449–1459. https://doi.org/10.1084/jem.169.4.1449
Panee J (2012) Monocyte chemoattractant protein 1 (MCP-1) in obesity and diabetes. Cytokine 60:1–12. https://doi.org/10.1016/j.cyto.2012.06.018
Van Coillie E, Van Damme J, Opdenakker G (1999) The MCP/eotaxin subfamily of CC chemokines. Cytokine Growth Factor Rev 10:61–86. https://doi.org/10.1016/s1359-6101(99)00005-2
Monteclaro FS, Charo IF (1997) The amino-terminal domain of CCR2 is both necessary and sufficient for high affinity binding of monocyte chemoattractant protein 1. Receptor activation by a pseudo- tethered ligand. J Biol Chem 272:23186–23190. https://doi.org/10.1074/jbc.272.37.23186
Cushing SD, Berliner JA, Valente AJ, Territo MC, Navab M, Parhami F, Gerrity R, Schwartz CJ, Fogelman AM (1990) Minimally modified low density lipoprotein induces monocyte chemotactic protein 1 in human endothelial cells and smooth muscle cells. Proc Natl Acad Sci USA 87:5134–5138. https://doi.org/10.1073/pnas.87.13.5134
Standiford TJ, Kunkel SL, Phan SH, Rollins BJ, Strieter RM (1991) Alveolar macrophage-derived cytokines induce monocyte chemoattractant protein-1 expression from human pulmonary type II-like epithelial cells. J Biol Chem 266:9912–9918. https://doi.org/10.1016/S0021-9258(18)92905-4
Sartipy P, Loskutoff DJ (2003) Monocyte chemoattractant protein 1 in obesity and insulin resistance. Proc Natl Acad Sci USA 100:7265–7270. https://doi.org/10.1073/pnas.1133870100
Wellen KE, Hotamisligil GS (2003) Obesity-induced inflammatory changes in adipose tissue. J Clin Investig 112:1785–1788. https://doi.org/10.1172/JCI20514
Daniele G, Guardado Mendoza R, Winnier D, Fiorentino TV, Pengou Z, Cornell J, Andreozzi F, Jenkinson C, Cersosimo E, Federici M, Tripathy D, Folli F (2014) The inflammatory status score including IL-6, TNF-α, osteopontin, fractalkine, MCP-1 and adiponectin underlies whole-body insulin resistance and hyperglycemia in type 2 diabetes mellitus. Acta Diabetol 51:123–131. https://doi.org/10.1007/s00592-013-0543-1
Nio Y, Yamauchi T, Iwabu M, Okada-Iwabu M, Funata M, Yamaguchi M, Ueki K, Kadowaki T (2012) Monocyte chemoattractant protein-1 (MCP-1) deficiency enhances alternatively activated M2 macrophages and ameliorates insulin resistance and fatty liver in lipoatrophic diabetic A-ZIP transgenic mice. Diabetologia 55:3350–3358. https://doi.org/10.1007/s00125-012-2710-2
Oh DY, Morinaga H, Talukdar S, Bae EJ, Olefsky JM (2012) Increased macrophage migration into adipose tissue in obese mice. Diabetes 61:346–354. https://doi.org/10.2337/db11-0860
Stefan N, Häring HU (2013) The role of hepatokines in metabolism. Nat Rev Endocrinol 9:144–152. https://doi.org/10.1038/nrendo.2012.258
Auberger P, Falquerho L, Contreres JO, Pages G, Le Cam G, Rossi B, Le Cam A (1989) Characterization of a natural inhibitor of the insulin receptor tyrosine kinase: cDNA cloning, purification, and anti-mitogenic activity. Cell 58:631–640. https://doi.org/10.1016/0092-8674(89)90098-6
Vionnet N, Hani EH, Dupont S, Gallina S, Francke S, Dotte S, De Matos F, Durand E, Leprêtre F, Lecoeur C, Gallina P, Zekiri L, Dina C, Froguel P (2000) Genomewide search for type 2 diabetes- susceptibility genes in French whites: evidence for a novel susceptibility locus for early-onset diabetes on chromosome 3q27-qter and independent replication of a type 2-diabetes locus on chromosome 1q21- q24. Am J Hum Genet 67:1470–1480. https://doi.org/10.1086/316887
Ishibashi A, Ikeda Y, Ohguro T, Kumon Y, Yamanaka S, Takata H, Inoue M, Suehiro T, Terada Y (2010) Serum fetuin-A is an independent marker of insulin resistance in Japanese men. J Atheroscler Thromb 17:925–933. https://doi.org/10.5551/jat.3830
Lin X, Braymer HD, Bray GA, York DA (1998) Differential expression of insulin receptor tyrosine kinase inhibitor (fetuin) gene in a model of diet-induced obesity. Life Sci 63:145–153. https://doi.org/10.1016/s0024-3205(98)00250-1
Stefan N, Hennige AM, Staiger H, Machann J, Schick F, Kröber SM, Machicao F, Fritsche A, Häring HU (2006) Alpha2-Heremans-Schmid glycoprotein/fetuin-A is associated with insulin resistance and fat accumulation in the liver in humans. Diabetes Care 29:853–857. https://doi.org/10.2337/diacare.29.04.06.dc05-1938
Dasgupta S, Bhattacharya S, Biswas A, Majumdar SS, Mukhopadhyay S, Ray S, Bhattacharya S (2010) NF-kappaB mediates lipid-induced fetuin-A expression in hepatocytes that impairs adipocyte function effecting insulin resistance. Biochem J 429:451–462. https://doi.org/10.1042/BJ20100330
Chatterjee P, Seal S, Mukherjee S, Kundu R, Mukherjee S, Ray S, Mukhopadhyay S, Majumdar SS, Bhattacharya S (2013) Adipocyte fetuin-A contributes to macrophage migration into adipose tissue and polarization of macrophages. J Biol Chem 288:28324–28330. https://doi.org/10.1074/jbc.C113.495473
Agarwal S, Chattopadhyay M, Mukherjee S, Dasgupta S, Mukhopadhyay S, Bhattacharya S (2017) Fetuin-A downregulates adiponectin through Wnt-PPARγ pathway in lipid induced inflamed adipocyte. Biochim Biophys Acta Mol Basis Dis 1863:174–181. https://doi.org/10.1016/j.bbadis.2016.10.002
Chattopadhyay M, Mukherjee S, Chatterjee SK, Chattopadhyay D, Das S, Majumdar SS, Mukhopadhyay S, Mukherjee S, Bhattarcharya S (2018) Impairment of energy sensors, SIRT1 and AMPK, in lipid induced inflamed adipocyte is regulated by Fetuin A. Cell Signal 42:67–76. https://doi.org/10.1016/j.cellsig.2017.10.005
Das S, Chattopadhyay D, Chatterjee SK, Mondal SA, Majumdar SS, Mukhopadhyay S, Saha N, Velayutham R, Bhattacharya S, Mukherjee S (2021) Increase in PPARγ inhibitory phosphorylation by Fetuin-A through the activation of Ras-MEK-ERK pathway causes insulin resistance. Biochim Biophys Acta Mol Basis Dis 1867:166050. https://doi.org/10.1016/j.bbadis.2020.166050
Chattopadhyay D, Das S, Guria S, Basu S, Mukherjee S (2021) Fetuin-A regulates adipose tissue macrophage content and activation in insulin resistant mice through MCP-1 and iNOS: Involvement of IFNγ-JAK2-STAT1 pathway. Biochem J 478:4027–4043. https://doi.org/10.1042/BCJ20210442
Mukhuty A, Fouzder C, Mukherjee S, Malick C, Mukhopadhyay S, Bhattacharya S, Kundu R (2017) Palmitate induced Fetuin-A secretion from pancreatic β-cells adversely affects its function and elicits inflammation. Biochem Biophys Res Commun 491:1118–1124. https://doi.org/10.1016/j.bbrc.2017.08.022
Nag S, Mandal S, Majumdar T, Mukhopadhyay S, Kundu R (2023) FFA-Fetuin-A regulates DPP-IV expression in pancreatic beta cells through TLR4-NFkB pathway. Biochem Biophys Res Commun 647:55–61. https://doi.org/10.1016/j.bbrc.2023.01.070
Kumar KG, Trevaskis JL, Lam DD, Sutton GM, Koza RA, Chouljenko VN, Kousoulas KG, Rogers PM, Kesterson RA, Thearle M, Ferrante AW Jr, Mynatt RL, Burris TP, Dong JZ, Halem HA, Culler MD, Heisler LK, Stephens JM, Butler AA (2008) Identification of adropin as a secreted factor linking dietary macronutrient intake with energy homeostasis and lipid metabolism. Cell Metab 8:468–481. https://doi.org/10.1016/j.cmet.2008.10.011
Butler AA, Tam CS, Stanhope KL, Wolfe BM, Ali MR, O’Keeffe M, St-Onge MP, Ravussin E, Havel PJ (2012) Low circulating adropin concentrations with obesity and aging correlate with risk factors for metabolic disease and increase after gastric bypass surgery in humans. J Clin Endocrinol Metab 97:3783–3791. https://doi.org/10.1210/jc.2012-2194
Chen X, Chen S, Shen T, Yang W, Chen Q, Zhang P, You Y, Sun X, Xu H, Tang Y, Mi J, Yang Y, Ling W (2020) Adropin regulates hepatic glucose production via PP2A/AMPK pathway in insulin- resistant hepatocytes. FASEB J 34:10056–10072. https://doi.org/10.1096/fj.202000115RR
Gao S, Ghoshal S, Zhang L, Stevens JR, McCommis KS, Finck BN, Lopaschuk GD, Butler AA (2019) The peptide hormone adropin regulates signal transduction pathways controlling hepatic glucose metabolism in a mouse model of diet-induced obesity. J Biol Chem 294:13366–13377. https://doi.org/10.1074/jbc.RA119.008967
Akcılar R, EmelKoçak F, Şimşek H, Akcılar A, Bayat Z, Ece E, Kökdaşgil H (2016) The effect of adropin on lipid and glucose metabolism in rats with hyperlipidemia. Iran J Basic Med Sci 19:245–251
Misu H, Takamura T, Takayama H, Hayashi H, Matsuzawa-Nagata N, Kurita S, Ishikura K, Ando H, Takeshita Y, Ota T, Sakurai M, Yamashita T, Mizukoshi E, Yamashita T, Honda M, Miyamoto K, Kubota T, Kubota N, Kadowaki T, Kim HJ, Lee IK, Minokoshi Y, Saito Y, Takahashi K, Yamada Y, Takakura N, Kaneko S (2010) A liver-derived secretory protein, selenoprotein P, causes insulin resistance. Cell Metab 12:483–495. https://doi.org/10.1016/j.cmet.2010.09.015
Saito Y, Hayashi T, Tanaka A, Watanabe Y, Suzuki M, Saito E, Takahashi K (1999) Selenoprotein P in human plasma as an extracellular phospholipid hydroperoxide glutathione peroxidase. Isolation and enzymatic characterization of human selenoprotein p. J Biol Chem 274:2866–2871. https://doi.org/10.1074/jbc.274.5.2866
Saito Y, Takahashi K (2002) Characterization of selenoprotein P as a selenium supply protein. Eur J Biochem 269:5746–5751. https://doi.org/10.1046/j.1432-1033.2002.03298.x
Satoh K (2021) Drug discovery focused on novel pathogenic proteins for pulmonary arterial hypertension. J Cardiol 78:1–11. https://doi.org/10.1016/j.jjcc.2021.01.009
Wallace IR, McKinley MC, Bell PM, Hunter SJ (2013) Sex hormone binding globulin and insulin resistance. Clin Endocrinol (Oxf) 78:321–329. https://doi.org/10.1111/cen.12086
Saez-Lopez C, Villena JA, Simó R, Selva DM (2020) Sex hormone-binding globulin overexpression protects against high-fat diet-induced obesity in transgenic male mice. J Nutr Biochem 85:108480. https://doi.org/10.1016/j.jnutbio.2020.108480
Simó R, Sáez-López C, Barbosa-Desongles A, Hernández C, Selva DM (2015) Novel insights in SHBG regulation and clinical implications. Trends Endocrinol Metab 26:376–383. https://doi.org/10.1016/j.tem.2015.05.001
Hara H, Yoshimura H, Uchida S, Toyoda Y, Aoki M, Sakai Y, Morimoto S, Shiokawa K (2001) Molecular cloning and functional expression analysis of a cDNA for human hepassocin, a liver-specific protein with hepatocyte mitogenic activity. Biochim Biophys Acta 1520:45–53. https://doi.org/10.1016/s0167-4781(01)00249-4
Wu HT, Ou HY, Hung HC, Su YC, Lu FH, Wu JS, Yang YC, Wu CL, Chang CJ (2016) A novel hepatokine, HFREP1, plays a crucial role in the development of insulin resistance and type 2 diabetes. Diabetologia 59:1732–1742. https://doi.org/10.1007/s00125-016-3991-7
Wu HT, Lu FH, Ou HY, Su YC, Hung HC, Wu JS, Yang YC, Wu CL, Chang CJ (2013) The role of hepassocin in the development of non-alcoholic fatty liver disease [published correction appears in J Hepatol. 2017 Feb; 66(2):468]. J Hepatol 59:1065–1072. https://doi.org/10.1016/j.jhep.2013.06.004
Kharitonenkov A, Shiyanova TL, Koester A, Ford AM, Micanovic R, Galbreath EJ, Sandusky GE, Hammond LJ, Moyers JS, Owens RA, Gromada J, Brozinick JT, Hawkins ED, Wroblewski VJ, Li DS, Mehrbod F, Jaskunas SR, Shanafelt AB (2005) FGF-21 as a novel metabolic regulator. J Clin Investig 115:1627–1635. https://doi.org/10.1172/JCI23606
Camporez JP, Jornayvaz FR, Petersen MC, Pesta D, Guigni BA, Serr J, Zhang D, Kahn M, Samuel VT, Jurczak MJ, Shulman GI (2013) Cellular mechanisms by which FGF21 improves insulin sensitivity in male mice. Endocrinology 154:3099–3109. https://doi.org/10.1210/en.2013-1191
Yu Y, He J, Li S, Song L, Guo X, Yao W, Zou D, Gao X, Liu Y, Bai F, Ren G, Li D (2016) Fibroblast growth factor 21 (FGF21) inhibits macrophage-mediated inflammation by activating Nrf2 and suppressing the NF-κB signaling pathway. Int Immunopharmacol 38:144–152. https://doi.org/10.1016/j.intimp.2016.05.026
McPherron AC, Lawler AM, Lee SJ (1997) Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387:83–90. https://doi.org/10.1038/387083a0
Consitt LA, Clark BC (2018) The vicious cycle of myostatin signaling in sarcopenic obesity: myostatin role in skeletal muscle growth, insulin signaling and implications for clinical trials. J Frailty Aging 7:21–27. https://doi.org/10.14283/jfa.2017.33
Guo T, Jou W, Chanturiya T, Portas J, Gavrilova O, McPherron AC (2009) Myostatin inhibition in muscle, but not adipose tissue, decreases fat mass and improves insulin sensitivity. PLoS ONE 4:e4937. https://doi.org/10.1371/journal.pone.0004937
Lin J, Arnold HB, Della-Fera MA, Azain MJ, Hartzell DL, Baile CA (2002) Myostatin knockout in mice increases myogenesis and decreases adipogenesis. Biochem Biophys Res Commun 291:701–706. https://doi.org/10.1006/bbrc.2002.6500
Zhang C, McFarlane C, Lokireddy S, Masuda S, Ge X, Gluckman PD, Sharma M, Kambadur R (2012) Inhibition of myostatin protects against diet-induced obesity by enhancing fatty acid oxidation and promoting a brown adipose phenotype in mice. Diabetologia 55:183–193. https://doi.org/10.1007/s00125-011-2304-4
Hu SL, Chang AC, Huang CC, Tsai CH, Lin CC, Tang CH (2017) Myostatin promotes interleukin-1β expression in rheumatoid arthritis synovial fibroblasts through inhibition of miR-21-5p. Front Immunol 8:1747. https://doi.org/10.3389/fimmu.2017.01747
Boström P, Wu J, Jedrychowski MP, Korde A, Ye L, Lo JC, Rasbach KA, Boström EA, Choi JH, Long JZ, Kajimura S, Zingaretti MC, Vind BF, Tu H, Cinti S, Højlund K, Gygi SP, Spiegelman BM (2012) A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 481:463–468. https://doi.org/10.1038/nature10777
Shan T, Liang X, Bi P, Kuang S (2013) Myostatin knockout drives browning of white adipose tissue through activating the AMPK-PGC1α-Fndc5 pathway in muscle. FASEB J 27:1981–1989. https://doi.org/10.1096/fj.12-225755
Huh JY, Mougios V, Kabasakalis A, Fatouros I, Siopi A, Douroudos II, Filippaios A, Panagiotou G, Park KH, Mantzoros CS (2014) Exercise-induced irisin secretion is independent of age or fitness level and increased irisin may directly modulate muscle metabolism through AMPK activation. J Clin Endocrinol Metab 99:E2154–E2161. https://doi.org/10.1210/jc.2014-1437
Liu TY, Shi CX, Gao R, Sun HJ, Xiong XQ, Ding L, Chen Q, Li YH, Wang JJ, Kang YM, Zhu GQ (2015) Irisin inhibits hepatic gluconeogenesis and increases glycogen synthesis via the PI3K/Akt pathway in type 2 diabetic mice and hepatocytes. Clin Sci 129:839–850. https://doi.org/10.1042/CS20150009
Tang H, Yu R, Liu S, Huwatibieke B, Li Z, Zhang W (2016) Irisin inhibits hepatic cholesterol synthesis via AMPK-SREBP2 signaling. EBioMedicine 6:139–148. https://doi.org/10.1016/j.ebiom.2016.02.041
Park MJ, Kim DI, Choi JH, Heo YR, Park SH (2015) New role of irisin in hepatocytes: the protective effect of hepatic steatosis in vitro. Cell Signal 27:1831–1839. https://doi.org/10.1016/j.cellsig.2015.04.010
Nishizawa H, Matsuda M, Yamada Y, Kawai K, Suzuki E, Makishima M, Kitamura T, Shimomura I (2004) Musclin, a novel skeletal muscle-derived secretory factor. J Biol Chem 279:19391–19395. https://doi.org/10.1074/jbc.C400066200
Chen WJ, Liu Y, Sui YB, Zhang B, Zhang XH, Yin XH (2017) Increased circulating levels of musclin in newly diagnosed type 2 diabetic patients. Diabetes Vasc Dis Res 14:116–121. https://doi.org/10.1177/1479164116675493
Acknowledgements
S. Guria, S. Basu, A. Hoory and S. Mukherjee gratefully acknowledge the Head, Department of Zoology (supported by DST-PURSE, Govt. of India), Visva-Bharati, Santiniketan for providing all infrastructural facilities. S. Basu and S. Mukherjee thank SERB, Govt. of India (No. ECR/2017/002470/LS). S. Guria is grateful to CSIR, Govt. of India for senior research fellowship (09/202(0098)/2019-EMR-I). A. Hoory acknowledges UGC, Govt. of India for junior research fellowship [F.82-1/2018(SA-III)].
Author information
Authors and Affiliations
Contributions
SG and SB: resources, visualization, writing—original draft; AH: resources, writing—original draft; SM and SM: conceptualization, resources, visualization, supervision, project administration, writing—original draft, writing—review and editing. All authors have read and approve the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no competing interests associated with the manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Guria, S., Basu, S., Hoory, A. et al. Inflammatory Overtones of Organokines in Metabolic Syndrome and Type 2 Diabetes. J Indian Inst Sci 103, 103–121 (2023). https://doi.org/10.1007/s41745-023-00391-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41745-023-00391-8