Abstract
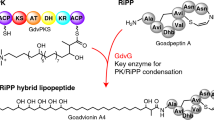
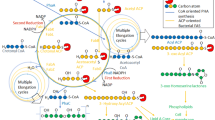
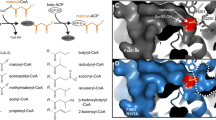
Fatty acyl-AMP ligases (FAAL) are enzymes that establish the crosstalk between fatty acid synthases and polyketide synthases or non-ribosomal peptide synthetases by activating newly synthesized fatty acids, shuttling them towards polyketide synthesis. These enzymes are unique as they have evolved a strict rejection mechanism for the coenzyme-A but can recognize and react with the phosphopantetheine of acyl-carrier proteins. A strategically placed insertion in the N-terminal domain and the rigidity of the hydrophobic anchor holding the insertion is at the heart of such a discrimination mechanism, the molecular details of which is yet to be clearly understood. The unique structural features of the insertion have been exploited to filter out FAALs from other members of the superfamily, in silico. Interestingly, several independent studies have characterized FAALs from different organisms such as Legionella, Myxococcus, Ralstonia, Pseudoalteromonas, Sorangium, etc. to name a few, laying emphasis on the usage of a FAAL-specific biochemistry in these organisms, particularly for the production of important bioactive molecules. These bioactive molecules help in improving the fitness of these systems to tide over the competition for nutritional resources in their local niche. Substrate specificity of these enzymes is another interesting aspect, which may hint at their potential roles in the cell. Various substrate-bound crystal structures have been used to identify the determinants of chain-length specificity and predict the preferences for different FAALs (both mycobacterial and non-mycobacterial). The fascinating details of the mechanistic variation of these enzymes and the molecular determinants that define the chain-length specificity have been discussed herewith.






Similar content being viewed by others
References
Schmelz S, Naismith JH (2009) Adenylate-forming enzymes. Curr Opin Struct Biol 19:666–671
Estrada P, Manandhar M, Dong SH, Deveryshetty J, Agarwal V, Cronan JE, Nair SK (2017) The pimeloyl-CoA synthetase BioW defines a new fold for adenylate-forming enzymes. Nat Chem Biol 13:668–674
Wang M, Moynie L, Harrison PJ, Kelly V, Piper A, Naismith JH, Campopiano DJ (2017) Using the pimeloyl-CoA synthetase adenylation fold to synthesize fatty acid thioesters. Nat Chem Biol 13:660–667
Harvey EN (1957) A history of luminescence from the earliest times until 1900. American Philosophical Society, Philadelphia
Green AA, McElroy WD (1956) Crystalline firefly luciferase. Biochim Biophys Acta 20:170–176
Koo JA, Schmidt SP, Schuster GB (1978) Bioluminescence of the firefly: key steps in the formation of the electronically excited state for model systems. Proc Natl Acad Sci USA 75:30–33
Schuster GB, Dixon B, Koo JY, Schmidt SP, Smith JP (1979) Chemical mechanisms of chemi- and bioluminescence. Reactions of high energy content organic compounds. Photochem Photobiol 30:17–26
The PyMOL Molecular Graphics, Version 1.8, Schrödinger LLC
Lipmann F (1944) Enzymatic Synthesis Of Acetyl Phosphate. J Biol Chem 155:55–70
Berg P (1956) Acyl adenylates; an enzymatic mechanism of acetate activation. J Biol Chem 222:991–1013
Bar-Tana J, Shapiro B (1964) Studies on palmitoyl-coenzyme A synthetase. Biochem J 93:533–538
Bar-Tana J, Rose G (1968) Studies on medium-chain fatty acyl-coenzyme A synthetase. Enzyme fraction I: mechanism of reaction and allosteric properties. Biochem J 109:275–282
Bar-Tana J, Rose G, Shapiro B (1968) Studies on medium-chain fatty acyl-coenzyme A synthetase. Purification and properties. Biochem J 109:269–274
Chang KH, Xiang H, Dunaway-Mariano D (1997) Acyl-adenylate motif of the acyl-adenylate/thioester-forming enzyme superfamily: a site-directed mutagenesis study with the Pseudomonas sp. strain CBS3 4-chlorobenzoate:coenzyme A ligase. Biochemistry 36:15650–15659
Marahiel MA, Stachelhaus T, Mootz HD (1997) Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev 97:2651–2674
Gulick AM (2009) Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem Biol 4:811–827
Hisanaga Y, Ago H, Nakagawa N, Hamada K, Ida K, Yamamoto M, Hori T, Arii Y, Sugahara M, Kuramitsu S, Yokoyama S, Miyano M (2004) Structural basis of the substrate-specific two-step catalysis of long chain fatty acyl-CoA synthetase dimer. J Biol Chem 279:31717–31726
Kochan G, Pilka ES, von Delft F, Oppermann U, Yue WW (2009) Structural snapshots for the conformation-dependent catalysis by human medium-chain acyl-coenzyme A synthetase ACSM2A. J Mol Biol 388:997–1008
Yonus H, Neumann P, Zimmermann S, May JJ, Marahiel MA, Stubbs MT (2008) Crystal structure of DltA. Implications for the reaction mechanism of non-ribosomal peptide synthetase adenylation domains. J Biol Chem 283:32484–32491
Conti E, Franks NP, Brick P (1996) Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure 4:287–298
Wu R, Reger AS, Lu X, Gulick AM, Dunaway-Mariano D (2009) The mechanism of domain alternation in the acyl-adenylate forming ligase superfamily member 4-chlorobenzoate: coenzyme A ligase. Biochemistry 48:4115–4125
Wu R, Cao J, Lu X, Reger AS, Gulick AM, Dunaway-Mariano D (2008) Mechanism of 4-chlorobenzoate:coenzyme A ligase catalysis. Biochemistry 47:8026–8039
Mitchell CA, Shi C, Aldrich CC, Gulick AM (2012) Structure of PA1221, a nonribosomal peptide synthetase containing adenylation and peptidyl carrier protein domains. Biochemistry 51:3252–3263
Sundlov JA, Fontaine DM, Southworth TL, Branchini BR, Gulick AM (2012) Crystal structure of firefly luciferase in a second catalytic conformation supports a domain alternation mechanism. Biochemistry 51:6493–6495
Trivedi OA, Arora P, Sridharan V, Tickoo R, Mohanty D, Gokhale RS (2004) Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature 428:441–445
Arora P, Goyal A, Natarajan VT, Rajakumara E, Verma P, Gupta R, Yousuf M, Trivedi OA, Mohanty D, Tyagi A, Sankaranarayanan R, Gokhale RS (2009) Mechanistic and functional insights into fatty acid activation in Mycobacterium tuberculosis. Nat Chem Biol 5:166–173
Yamada KD, Tomii K, Katoh K (2016) Application of the MAFFT sequence alignment program to large data—reexamination of the usefulness of chained guide trees. Bioinformatics 32:3246–3251
Robert X, Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324
Hayashi T, Kitamura Y, Funa N, Ohnishi Y, Horinouchi S (2011) Fatty acyl-AMP ligase involvement in the production of alkylresorcylic acid by a Myxococcus xanthus type III polyketide synthase. ChemBioChem 12:2166–2176
Kage H, Kreutzer MF, Wackler B, Hoffmeister D, Nett M (2013) An iterative type I polyketide synthase initiates the biosynthesis of the antimycoplasma agent micacocidin. Chem Biol 20:764–771
Hemmerling F, Lebe K, Wunderlich J, Hahn F (2018) An unusual FAAL-ACP didomain in ambruticin biosynthesis. ChemBioChem 19:1006–1011
Marchetti PM, Kelly V, Simpson JP, Ward M, Campopiano DJ (2018) The carbon chain-selective adenylation enzyme TamA: the missing link between fatty acid and pyrrole natural product biosynthesis. Org Biomol Chem 16:2735–2740
Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797
Zhang Z, Zhou R, Sauder JM, Tonge PJ, Burley SK, Swaminathan S (2011) Structural and functional studies of fatty acyl adenylate ligases from E. coli and L. pneumophila. J Mol Biol 406:313–324
Goyal A, Verma P, Anandhakrishnan M, Gokhale RS, Sankaranarayanan R (2012) Molecular basis of the functional divergence of fatty acyl-AMP ligase biosynthetic enzymes of Mycobacterium tuberculosis. J Mol Biol 416:221–238
Gavalda S, Leger M, van der Rest B, Stella A, Bardou F, Montrozier H, Chalut C, Burlet-Schiltz O, Marrakchi H, Daffe M, Quemard A (2009) The Pks13/FadD32 crosstalk for the biosynthesis of mycolic acids in Mycobacterium tuberculosis. J Biol Chem 284:19255–19264
Trivedi OA, Arora P, Vats A, Ansari MZ, Tickoo R, Sridharan V, Mohanty D, Gokhale RS (2005) Dissecting the mechanism and assembly of a complex virulence mycobacterial lipid. Mol Cell 17:631–643
Siméone R, Léger M, Constant P, Malaga W, Marrakchi H, Daffe M, Guilhot C, Chalut C (2010) Delineation of the roles of FadD22, FadD26 and FadD29 in the biosynthesis of phthiocerol dimycocerosates and related compounds in Mycobacterium tuberculosis. FEBS J 277:2715–2725
Vergnolle O, Chavadi SS, Edupuganti UR, Mohandas P, Chan C, Zeng J, Kopylov M, Angelo NG, Warren JD, Soll CE, Quadri LE (2015) Biosynthesis of cell envelope-associated phenolic glycolipids in Mycobacterium marinum. J Bacteriol 197:1040–1050
Srivastava S, Chaudhary S, Thukral L, Shi C, Gupta RD, Gupta R, Priyadarshan K, Vats A, Haque AS, Sankaranarayanan R, Natarajan VT, Sharma R, Aldrich CC, Gokhale RS (2015) Unsaturated lipid assimilation by mycobacteria requires auxiliary cis-trans enoyl CoA isomerase. Chem Biol 22:1577–1587
Toyoda K, Inui M (2018) Extracytoplasmic function sigma factor sigma(D) confers resistance to environmental stress by enhancing mycolate synthesis and modifying peptidoglycan structures in Corynebacterium glutamicum. Mol Microbiol 107:312–329
Conti E, Stachelhaus T, Marahiel MA, Brick P (1997) Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J 16:4174–4183
Stachelhaus T, Mootz HD, Marahiel MA (1999) The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol 6:493–505
Khurana P, Gokhale RS, Mohanty D (2010) Genome scale prediction of substrate specificity for acyl adenylate superfamily of enzymes based on active site residue profiles. BMC Bioinform 11:57
Sankaranarayanan R, Saxena P, Marathe UB, Gokhale RS, Shanmugam VM, Rukmini R (2004) A novel tunnel in mycobacterial type III polyketide synthase reveals the structural basis for generating diverse metabolites. Nat Struct Mol Biol 11:894–900
Andersson CS, Lundgren CA, Magnúsdóttir A, Ge C, Wieslander A, Martinez Molina D, Hogbom M (2012) The Mycobacterium tuberculosis very-long-chain fatty acyl-CoA synthetase: structural basis for housing lipid substrates longer than the enzyme. Structure 20:1062–1070
Linne U, Schafer A, Stubbs MT, Marahiel MA (2007) Aminoacyl-coenzyme A synthesis catalyzed by adenylation domains. FEBS Lett 581:905–910
Marahiel MA (2016) A structural model for multimodular NRPS assembly lines. Nat Prod Rep 33:136–140
Kries H, Niquille DL, Hilvert D (2015) A subdomain swap strategy for reengineering nonribosomal peptides. Chem Biol 22:640–648
Acknowledgements
R. S. acknowledges funding from JC Bose Fellowship of Science and Engineering Research Board (SERB), India, and Centre of Excellence Project of Department of Biotechnology, India.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Priyadarshan, K., Sankaranarayanan, R. Fatty Acyl-AMP Ligases as Mechanistic Variants of ANL Superfamily and Molecular Determinants Dictating Substrate Specificities. J Indian Inst Sci 98, 261–272 (2018). https://doi.org/10.1007/s41745-018-0084-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41745-018-0084-2