Abstract
miR-18a has been identified as a significantly expressed microRNA (miRNA) in non-small cell lung cancer (NSCLC) and plays a vital role in cancer cell transformation, metastasis, and carcinogenesis. Herein, a pair of binary probes from numerous probe pairs based on single nucleotide polymorphism analyses of miR-18a and miR-18b was first designed and screened to develop a Y-shaped ratio biosensor for accurate detection of serum miR-18a in NSCLC. The special structure of the binary probes combined with the hairpin showed strong specificity for miR-18a, which was confirmed by polyacrylamide gel electrophoresis assay and square wave voltammetry assay. Furthermore, it is beneficial to immobilize single-stranded DNA (ssDNA) probes due to the large specific surface area of nanoporous gold, thereby improving the sensitivity of the biosensor. The Y-shaped ratio biosensor exhibited a wide detection range and can quantify the concentration of miR-18a in the range of 10 fmol/L–100 pmol/L, with a limit of detection of 0.211 fmol/L (S/N = 3). Moreover, it exhibits excellent detection capabilities in serum samples since the biosensor showed a high selectivity toward the coexistence of miR-18a and miR-18b. Therefore, the prepared Y-shaped ratio biosensor is a highly sensitive and specific miR-18a detection tool, capable of identifying microscopic amounts of miR-18a in serum samples, providing great potential for early screening of NSCLC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Early screening is critical to the treatment and prognosis of lung cancer [1]. Previous studies [2, 3] have shown that miR-18a is significantly increased in cells and peripheral blood of non-small cell lung cancer (NSCLC) patients. Therefore, miR-18a has become a crucial biomarker for the initiation of NSCLC. miR-18a is a member of the miR-17-92a cluster, and its transcription locus is in the 13q31.3 region of the human chromosome 13. miR-18b belongs to the miR-106a-363 cluster, which is the result of X chromosome transcription. The functions of miR-18b and miR-18a are very different, but they only differ by a single base. A study has pointed out that miR-18a and miR-18b had opposite expression results in colorectal cancer [4]. Moreover, Zhou et al. found that miR-18b was significantly downregulated in NSCLC [5]. Currently, there are extensive studies on the expression of miR-18a in the blood of lung cancer patients, but there is a lack of research on the expression of miR-18b [6]. The interference of miR-18b needs to be excluded during the detection of miR-18a.
There are few investigations on electrochemical biosensors for miR-18a. A biosensor for detecting miR-18a was constructed using the highly specific duplex cell-specific nuclease (DSN), which showed excellent selectivity and simultaneously achieved duplex-specific nuclease-assisted target recovery and miR-18a recycling [7]. The introduction of DSN amplifies the signal of the biosensor and improves the overall performance. Next, specific toehold-mediated strand displacement (TMSD) in conjunction with rolled-circle amplification reaction (RCA) mediated was utilized by Bst DNA polymerase, CircLigaseII ssDNA ligase, Exo-I, and Exo-III to collaboratively construct an electrochemical biosensor [8]. These structures enable it to provide a limit of detection (LOD) of 9 amol/L and high selectivity. However, miR-18b was not detected in their selective experiment. MiR-18b and miR-18a differ by a single base at the 3' end, making it more difficult to identify than the intermediate site [9]. In addition, both methods utilized biological enzymes and probe cycling, which was time-consuming. Furthermore, the use of enzymes had its drawbacks, including the impact of temperature, pH, cleanliness of the electrode surface, and other factors that could potentially destabilize the enzyme on the electrode [10]. We aim to create a biosensor that can detect miR-18a and differentiate miR-18b without using enzymes or chain amplification. The biosensor should be straightforward and quick, making it ideal for fast detection in clinical settings. Using binary probes to construct a Y-shaped biosensor, we can meet these above requirements and achieve selective and rapid detection of miR-18a [11]. Many studies have been conducted using the Y-shaped biosensor owing to its simple direct operation, and time saving. The first electrochemical biosensor with a Y-shaped structure to detect E-DNA was developed [12]. Their CP was without any redox reporter labeling and the biosensor just detected an electrochemical signal from AP with Fc attached close to the electrode surface. Although, their biosensor assay showed high sensitivity and specificity. The biosensor was challenging to achieve quality control because of the errors of CP solid-loading on gold electrodes and hybridization between probes, which only shows up in electrochemical signals and lacks experimental validation techniques such as electrophoresis. Therefore, subsequent researchers [13] built on this foundation to design ratio biosensors with Y-shaped for the detection of miR-21 from tumor cells. The biosensor was designed to trigger two hairpin structures to generate a chain hybridization reaction (CHA), and the resulting double-stranded body further binds to another hairpin assembled on the surface of the sensor to generate a stable Y-shaped DNA complex. Although the linear range reached 5 fmol/L—0.1 nmol/L and 1.1 fmol/L LOD. The total time taken from the start of miR-21 to trigger the CHA to the resultant SWV assay was about 3.5 h. Moreover, new errors may be introduced during the amplification of CHA.
The sensitivity of biosensors with Y-shaped structures may be limited due to the balance of high specificity in the construction, also. Moreover, the complexity of the components in peripheral blood as well as the low abundance of miR-18a requires high sensitivity. The main methods to increase the sensitivity of sensors are enzyme assistance [14], probe amplification [15], the application of nanomaterials [16], etc. Therefore, nanomaterials may be a better method to amplify the signals of biosensors. The metallic nanoporous materials can be widely used in the fields of catalysis of redox reactions, sensors, and the isolation of biomolecules due to their surface and structural properties [17]. Noble metal porous materials have been widely used in various aspects such as catalysis, biosensing technology, transmission media, porous electrodes, and energy storage [18]. Among these noble metals, the porous gold film has excellent properties such as good chemical stability and outstanding ductility. In particular, nanoporous gold (NPG) stands out as a widely applicable material due to its ease of preparation, better conductivity [19], and ductility compared to gold nanoparticles (AuNPs). The porous structure of NPG provides a higher specific surface area, smaller relative density, and improved adsorption capacity, creating more binding sites for biomolecular recognition elements and enhancing electrocatalytic ability [20]. As an immobilized carrier material, NPG shows great promise in constructing electrochemical biosensors for miRNA detection and may outperform other methods in mediated electrochemical biosensors [21].
In this work, a pair of binary probes was designed and screened by NUPACK (https://www.nupack.org/partition/new) and polyacrylamide gel electrophoresis (PAGE) from a large number of probe pairs to construct a Y-shaped ratio electrochemical biosensor for detecting miR-18a in peripheral blood with enhanced specificity. Then the NPG was introduced to improve the sensitivity of the biosensor. The prepared biosensor, which has a simple structure and does not require enzymes, antibodies, or chain amplification, can detect rapidly at room temperature with high specificity, sensitivity, and stability, and provide a new strategy for early screening of NSCLC.
2 Experimental
2.1 Chemicals, Reagents, and Apparatus
All chemically synthesized ssDNA strands and miRNAs were purchased from China Peptides Co., Ltd. (Wuhan, China), and their base sequences are shown as follows: target probe a (TPa, miR-18a): 5’-UAAGGUGCAUCUAGUGCAGAUAG-3’; target probe b (TPb, miR-18b): 5’-UAAGGUGCAUCUAGUGCAGUUAG-3’; auxiliary probe (AP) with methylene blue (Mb): 5’-Mb-GGATACCGGCTAGATGCACCTTA-3’; capture probe (CP) with ferrocene (Fc) and sulfhydryl (SH): 5’-Fc-GGATACTATCTGCACCGGTATCC-(CH2)6-SH-3’. All probes were dissolved and diluted to 100 μmol/L with RNase-free water. Ammonium persulfate (APS), N, N, N’, N’-tetramethylethylenediamine (TEMED), Bis-acrylamide (30%), GelRed nucleic acid dye, potassium ferricyanide (K3Fe(CN)6), potassium ferrocyanide trihydrate (K4Fe(CN)6), ethylenediaminetetraacetic acid (EDTA), RNase-free water, KCl, and NaCl, were purchased from Shanghai Macklin Biochemical Co., Ltd. (Shanghai, China). Tris-(2-carboxyethyl) phosphine hydrochloride (TCEP), 6-mercaptoethanol (MCH), and 0.1 mol/L phosphate buffer solution (PBS: 1.37 mmol/L NaCl, 26.8 mmol/L KCl, 81.0 mmol/L Na2HPO4, 17.6 mmol/L KH2PO4, pH 7.4) were purchased from Sigma-Aldrich (U.S.). 12 K Au–Ag alloy films were purchased from Nanjing Yongbo Metal Materials Co., Ltd. (Nanjing, China).
CHI Instruments Model 760B electrochemical analyzer (CHI Inc., Shanghai, China) was used. All solutions were prepared with Milli-Q water (resistivity of 18 MΩ·cm) from a Millipore ultrapure water purification system.
2.2 Gel Electrophoresis
To determine the efficacy of the proposed biosensing strategy, 15% native PAGE was carried out to analyze probe products that hybridized at room temperature for 60 min. Each lane was loaded with 5 μL of sample and ran in 1 × TBE buffer at 100 V for about 60 min. Upon completion, the gel was stained by GelRed nuclear staining for 5 min and scanned using a Bio-Red Imaging System.
2.3 Bare Gold Electrode (Au-E) Pretreatment
The Au-E was carefully polished to smooth with alumina slurry (1.0, 0.3, and 0.05 µm) in sequence, followed by ultrasonication in 50 wt % HNO3, ethanol, and Milli-Q water for 1 min. Thereafter, Au-E was dried with infrared lamps.
2.4 Synthesis of NPG by Dealloying
NPG was prepared by dealloying Au–Ag alloy thin film in concentrated nitric acid (HNO3). First, 12 k Au–Ag alloy thin film was floated in concentrated HNO3 (30 °C, 65 wt %) for 120 min. Then any excess acid was removed by HNO3 and ultrapure water washing. Finally, NPG was adhered to the surface of the polished and cleaned Au-E, and dried by infrared lamp.
2.5 Preparation of the Biosensor
To reduce the disulfide bonds, 2 μL of CP (10 μmol/L), 1 μL of TCEP (10 mmol/L), and 7 μL of hybridization buffer were mixed for 1 h. Subsequently, 5 μL of mixed solution was added dropwise on the surface of the Au/NPG electrode and incubated overnight to produce the Au/NPG-CP electrode. Thereafter, the Au/NPG-CP electrode was immersed in 1 mmol/L MCH solution for 60 min after being gently washed with 10 mmol/L PBS (pH 7.4). Thus, the Au/NPG-CP/MCH electrode was obtained. All the processes were completed at room temperature.
2.6 Detection of miR-18a(TPa) by the Biosensor
5 μL of mixed solution containing AP (2 μmol/L) and TPa was added to the surface of the Au/NPG-CP/MCH electrode and incubated at room temperature for 80 min. The electrochemical performance of the biosensor was investigated by square wave voltammetry (SWV) from −0.5 V to 0.6 V in 10 mmol/L PBS (pH 7.4) solution, with an amplitude of 50 V, and a frequency of 100 Hz. Electrochemical impedance spectroscopy (EIS) and cyclic voltammetry (CV) were processed in ferricyanide solution (5.0 mmol/L [Fe(CN)6]3−/4− and 0.1 mol/L KCl). Scanning potential and scanning rate of CV were from − 0.2 V to 0.6 V and 0.05 V/s, respectively. EIS was carried out with a frequency from 0.1 Hz to 100 kHz. The scanned SWV, CV, and EIS curves were analyzed using ORIGIN 2021.
2.7 Recovery Test of miR-18a in Serum Samples
Human serum was extracted from healthy human whole blood sample by centrifuging at 3000 r/min for 10 min. Healthy human serum and de-complemented fetal bovine serum (FBS) were diluted twofold and tenfold with 10 mmol/L PBS (pH 7.4), heated at 95 °C for 15 min, ice bathed for 2 min, and centrifuged at 12,000 r/min for 2 min to detect miR-18a content. To further evaluate the effectiveness of the electrode response to miR-18a, recovery experiments were conducted. Serum samples with miR-18a concentrations of 10 pmol/L, 1 pmol/L, and 100 fmol/L were prepared and assayed. Three samples were detected for each concentration and the signal changes were the average of three results.
3 Results and Discussion
3.1 Design Principle of the Biosensing
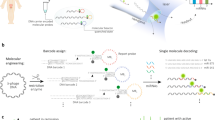
The NPG-mediated Y-shaped ratio biosensor is as illustrated in Scheme 1. The biosensor hybridizes by opening the hairpin structure of the CP in the presence of both AP and TPa. CP-AP-TPa hybridization product with a Y-shaped structure was constructed. The current signals of the Fc and Mb will change through the conformational change of CP and the introduction of AP, since CP and AP are connected to the redox reporter Fc and Mb, respectively. Then the TPa concentration is calculated based on the change of IMb/IFc ratio caused by the change in the Mb and Fc current signals before and after hybridization, which reflects the hybridization efficiency. It is worth emphasizing that the increase in the spatial distance between probes has little effect on the specific hybridization efficiency as the TPa concentration decreases within a certain range [22].
3.2 Characterization of NPG
Scanning electron microscopy (SEM, TESCAN MIRA LMS, Czech Republic) was used to study the nano- and micrometer-scale morphological features by photographing the samples at 50 k × and 100 k × magnifications (Fig. 1a, b). The pore morphology of the NPG is the most important feature that affects the electrochemical performance. As shown in Fig. 1a, b, the prepared NPG can be seen in the entire field and the three-dimensional nanoporous structure distributed uniformly with a pore size about 10 to 100 nm, which can provide a large number of binding sites for the CP probe and amplify the signal of the sensor.
3.3 Characterization of the Biosensor Fabrication
The stepwise fabrication of the biosensor was demonstrated by the results of EIS and CV (Fig. 1c, d). The bare gold electrode (bare Au-E) displayed a very small semicircle of EIS signal, where the semicircle diameter equals to the interfacial electron transfer resistance (Ret), as well as a few reversible [Fe (CN) 6]3−/4− redox peaks (curve 1). Compared to the bare Au-E, the Ret and redox intensity of the NPG-modified electrode (Au/NPG) were significantly reduced (curve 2), which was attributed to the large surface area and high conductivity of NPG. When the capture probe CP was assembled onto the electrode surface (Au/NPG-CP), electron transfer was greatly hindered by the negative charge of the DNA [23], increasing the Ret and redox intensity (curve 3). Subsequently, the non-conducting MCH coated on the electrode (Au/NPG-CP-MCH) blocked the active sites on the electrode surface, effectively covering the uncovered space and causing Ret to continue increasing (curve 4). As AP and TPa further hybridized on the electrode (Au/NPG-CP-MCH-AP-TPa), the electrode surface became more negatively charged, leading to a further increase in Ret and redox intensity (curve 5). The results indicated the successful construction of the Y-shaped biosensor.
3.4 Feasibility Analysis
PAGE was used to analyze the feasibility of probe hybridization (Fig. 2a). Lanes 1–4 show the molecular weights and structures of CP, AP, TPa, and TPb. There is no hybridization between CP and AP, TPa, or TPb (lanes 5–7), and there are hybridizations between AP and TPa or TPb (lanes 8 and 9). The result of the three-probe hybridization system indicated that CP-AP-TPa successfully hybridized in lane 10, but CP and AP-TPb did not exhibit hybridization (lane 11). The results of PAGE indicated that the CP-AP-TPa hybridization had high specificity. However, we can judge from the brightness of the electrophoretic band in lane 10 that the hybridization efficiency of CP, AP, and TPa at a concentration of 1 μmol/L was insufficient.
This hybridization property was achieved by selection of complementary pairing sequences between AP, CP, and TPa. AP and CP were designed to have a 9-base complementary pairing region (-GGATACCGG-), with a melting temperature of approximately 30 °C. The hybridization temperature was set below 25 °C. TPa and CP were paired with a 9-base complementary pairing region (–CTATCTGCA–) at a hybridization temperature of about 21 °C. The CP hairpin structure with a free energy of −2.5 kcal/mol according to John’s calculations [24] was stable. A temperature about 50 °C was required for opening this hairpin structure, which was significantly higher than that of the hybridization between AP or TPa and CP. Therefore, at room temperature, neither AP nor TPa alone was able to open the hairpin structure of CP for hybridization. The experimental results, especially the PAGE analysis of the probe hybridization, aligned with the design expectations. This indicated that the AP and CP sequences can meet assay requirements of specificity, sensitivity, and false-positive prevention.
The SWV detection was used to verify the feasibility of this Y-shaped biosensor (Fig. 2b). In similar conditions, this Y-shaped biosensor with Au/NPG was able to detect larger Fc and Mb signals than the bare Au-E at room temperature. In the presence of only CP, a strong Fc signal but no Mb signal was detected due to the hairpin structure that brought Fc close to the electrode surface. However, in the absence of TPa, the CP hairpin structure was difficult to be opened by AP, resulting in a high Fc signal and a weak Mb signal from non-specific adsorption of probe AP. The CP hairpin structure was easily transformed into the Y-shaped structure by hybridization when both AP and TPa existed. The alteration in probe shape caused the Fc to move away from the electrode surface while Mb to move closer, resulting in a weaker Fc signal and a higher Mb signal. When TPb and AP were both present, CP cannot fully hybridize with TPb, making it difficult to open the hairpin structure of CP by AP-TPb. As a result, the changes in the Fc and Mb signals were relatively weak. The tests using only 100 pmol/L TPa and mixing with equal concentration of TPb showed nearly identical results, proving the biosensor’s exceptional specificity. The above results validated the feasibility of the present Y-shaped electrochemical biosensor for target detection.
3.5 Optimization of the Biosensing Conditions
In Y-shaped biosensors, factors like CP concentration and hybridization time can affect the experimental results. We used 100 pmol/L of TPa and 2 μmol/L of AP to detect the signals of CP with different concentrations, and the response of IMb/IFc changed with different CP concentration. Since a weak Mb signal was still detected in the presence of AP only, we optimized the CP concentration based on the signal-to-noise ratio (IMb/IFc)(AP-TPa)/(IMb/IFc)AP. The results in Fig. 3a indicated that the signal-to-noise ratio of the system increased rapidly at the CP concentrations of up to 2 μmol/L, but decreased beyond that, so the CP concentration of 2 μmol/L was chosen for subsequent experiments. As shown in Fig. 3b, at CP concentration of 2 μmol/L, the value of IMB/IFc significantly rises with longer incubation time of up to 80 min.
Optimizations of experimental parameters: a concentration of CP probes and b incubation time of surface hybridization. c The linear range, d selectivity, e repeatability, and f stability of the biosensor. (Error bars represent the standard deviations measured from three different tests. Scan range from −0.5 to 0.6 V, amplitude 0.05 V, frequency 100 Hz in 10 mmol/L PBS buffer.)
3.6 Sensitivity, Selectivity, Repeatability, and Stability Analysis of Y-Shaped Biosensor
Under the optimal conditions, different concentrations of TPa were detected by SWV. As the concentration of TPa increased, the current IMb became larger while the IFc became smaller. The linear regression equation is IMb/IFc = 0.06213lgc(TPa) + 0.05629, with the concentration of TPa ranging from 10 fmol/L to 100 pmol/L (Fig. 3c). The correlation coefficient was 0.9982, and the LOD was 0.211 fmol/L (S/N = 3).
To ascertain the selectivity of the Y-shaped electrochemical biosensor, the concentrations of TPa and TPb were maintained at 1 nmol/L, a level above the detection limit. The achieved data indicated that the difference of IMb/IFc between TPa and TPb was more than five times (Fig. 3d). The results demonstrated strong selectivity in separating well-matched targets from mismatched target sequences, which aligns with the outcomes seen in the PAGE analysis (Fig. 2a). This is because the Y-shaped structure reduces the binding rates of mismatched targets, while the CP hairpin structure can only be opened by the precisely matched TPa. Several studies [25, 26] on single nucleotide polymorphism (SNP) have proposed that hairpin structures unite binary DNA probe molecular beacons (also known as binary probes) with the shorter probe binding to the mismatched base. Both probes hybridize only when they bind to the target, significantly enhancing the sensitivity and specificity of SNP detection. This study demonstrated that the Y-shaped biosensor has excellent selectivity for recognizing sequences with single base mismatches. In the case of mixing with same concentration of miR-18b, the signal intensity was almost the same as that of detecting only miR-18a. This anti-interference ability is crucial for clinical applications, as serum samples contain numerous interferences.
In addition, we analyzed the results of 5 different batches of biosensors for detecting 100 pmol/L of TPa. The relative standard deviation (RSD) was 4.6%, proving that the electrodes have good repeatability between batches (Fig. 3e).
Furthermore, the biosensor’s stability was evaluated. As shown in Fig. 3f, Au/NPG-CP/MCH electrodes were stored at 4 °C and measured miR-18a every 5 d. After 15 d, the biosensor response still kept up to 95.59% of the initial. This indicated that the biosensor was stable, which can increase its lifespan. These features are beneficial in clinical assays, as the biosensor is likely to perform with adequate power.
3.7 Recovery Test of miR-18a in Diluted Serum Samples
The Y-shaped biosensor successfully detected miR-18a levels of about 24.165 fmol/L and 121.997 fmol/L in tenfold and twofold of healthy human serum, but not in FBS. miR-18a levels in healthy human serum samples were calculated to be about 242.823 fmol/L, which is almost identical to the data of Hirajima et al. [27], which were between 0.19 and 1.27 amol/μL, with a mean value of 0.73 amol/μL. Using qRT-PCR combined with a standard curve was able to measure the levels of miR-18a in the serum of patients with esophageal squamous carcinoma and healthy individuals. In this study, the process of detecting miR-18a in serum samples only requires dilution of serum samples and heating at 95 °C for 15 min [28, 29], without separation, extraction, and amplification. miR-18a in the blood is present in exosome vesicles, and high-temperature cleavage of the vesicles releases the miR-18a so that it can be detected by the biosensor. Moreover, the protein in serum can be denatured after high temperature and the interference of protein components can be removed completely by centrifugation, which makes the detection of miRNA in serum more easy.
To further evaluate the efficiency of the biosensor response to miR-18a in real samples, the tenfold serum samples with miR-18a concentrations of 10 pmol/L, 1 pmol/L, and 100 fmol/L were prepared and measured (Table 1). Each concentration was examined three times. The amount of miR-18a in serum was subtracted from the detected result and divided by the added concentration to obtain the recovery. The results showed the recovery ranged from 95.41% to 104.81% with the RSD of 1.81%–4.01%. This indicated the proposed Y-shaped biosensor can accurately detect miR-18a in serum samples.
4 Conclusion
In this work, a pair of binary probes was designed and screened to develop a Y-shaped ratiometric biosensor for accurate detection of miR-18a of NSCLC serum. The design of the binary probes combined with the hairpin structure is highly specific in identifying miR-18a. NPG was synthesized to provide a larger surface for ssDNA sequestration, which significantly improved the sensitivity of the biosensor with a linear range of 10 fmol/L-100 pmol/L and a LOD of 0.211 fmol/L (S/N = 3). The biosensor is easy to operate, no need of enzyme, antibody, chain amplification, and extension, and can realize rapid and stable detection with strong specificity and high sensitivity at room temperature. Moreover, it can be applied to detection in human serum. This simple and cost-effective assay is more suitable for early screening in the clinic, which provides a broad prospect for the clinical diagnosis and treatment of early NSCLC.
Data availability
All data that support the findings of this study are included within the article.
References
Li W, Liu J-B, Hou L-K, Yu F, Zhang J, Wu W, Tang X-M, Sun F, Lu H-M, Deng J, et al. Liquid biopsy in lung cancer: significance in diagnostics, prediction, and treatment monitoring. Mol Cancer. 2022;21(1):1–15.
Komatsu S, Ichikawa D, Takeshita H, Morimura R, Hirajima S, Tsujiura M, Kawaguchi T, Miyamae M, Nagata H, Konishi H. Circulating miR-18a: a sensitive cancer screening biomarker in human cancer. In Vivo. 2014;28:293–7.
Shen K, Cao Z, Zhu R, You L, Zhang T. The dual functional role of MicroRNA-18a (miR-18a) in cancer development. Clin Transl Med. 2019;8:32–44.
Dai J, Li X, Xia J, Jin H. Expression and clinical significance of miR-18a and miR-18b in colon cancer. Jiangsu Med. 2011;37:2284–6.
Zhou C, Chen Z, Zhao L, Zhao W, Zhu Y, Liu J, Zhao X. A novel circulating miRNA-based signature for the early diagnosis and prognosis prediction of non–small-cell lung cancer. J Clin Lab Anal. 2020;34(11): e23505.
Shao L, Lu X, Zhou Y, Wang Y, Wang X, Zhuang Z, Gong J. Altered miR-93-5p/miR-18a expression in serum for diagnosing non-small cell lung cancer. Am J Trans Res. 2021;13:5073–9.
Rezaei H, Hosseini M, Radfar S. A dual-signaling electrochemical ratiometric strategy combining “signal-off” and “signal-on” approaches for detection of MicroRNAs. Anal Biochem. 2021;632: 114356.
Niazi M, Azizi A, Khajavi Z, Sheikh M, Taheri S, Radfar S, Alizadeh A, Ghanbari R. A universal ratiometric method for Micro-RNA detection based on the ratio of electrochemical/electrochemiluminescence signal, and toehold-mediated strand displacement amplification. Anal Chim Acta. 2023;1257: 341119.
Milkani E, Morais S, Lambert CR, McGimpsey WG. Detection of oligonucleotide systematic mismatches with a surface plasmon resonance sensor. Biosens Bioelectron. 2010;25:1217–20.
Eslami R, Azizi N, Ghaffarian SR, Mehrvar M, Zarrin H. Highly sensitive and selective non-enzymatic measurement of glucose using arraying of two separate sweat sensors at physiological pH. Electrochim Acta. 2022;404: 139749.
Hosseini SS, Jebelli A, Vandghanooni S, Jahanban-Esfahlan A, Baradaran B, Amini M, Bidar N, de la Guardia M, Mokhtarzadeh A, Eskandani M. Perspectives and trends in advanced DNA biosensors for the recognition of single nucleotide polymorphisms. Chem Eng J. 2022;441: 135988.
Jia L-P, Zhao R-J, Feng Z, Wang M-Y, Ma R-N, Jia W-L, Shang L, Zhang W, Xue Q-W, Wang H-S. Ultrasensitive electrochemical detection of circulating tumor DNA by hollow polymeric nanospheres and dual enzyme assisted target amplification strategy. Sens Actuators, B Chem. 2022;350: 130849.
Xue Y, Xie H, Wang Y, Feng S, Sun J, Huang J, Yang X. Novel and sensitive electrochemical/fluorescent dual-mode biosensing platform based on the cascaded cyclic amplification of enzyme-free DDSA and functional nucleic acids. Biosens Bioelectron. 2022;218: 114762.
Campuzano S, Pedrero M, Barderas R, Pingarrón JM. Empowering electrochemical biosensing through nanostructured or multifunctional nucleic acid or peptide biomaterials. Adv Mater Technol. 2022;7:2200310.
Cao Y, Ma C, Zhu JJ. DNA technology-assisted signal amplification strategies in electrochemiluminescence bioanalysis. J Anal Test. 2021;5(2):95–111.
Wang XY, Li Y, Lv S, Bi S. pH-responsive magnetic I-motif container coupled with DNA walker for construction of dual-signal electrochemical biosensor. J Anal Test. 2022;6(1):12–9.
Xu W, Cui Z, Zhu S. Recent advances in open-cell porous metal materials for electrocatalytic and biomedical applications. Acta Metall Sin. 2022;58(12):1527–44.
Xiao S, Wang S, Wang X, Xu P. Nanoporous gold: a review and potentials in biotechnological and biomedical applications. Nano Select. 2021;2:1437–58.
Xu Y, Wang X, Chen H, Chen L, Chen W, Yin X, Liu A, Lin X, Weng S, Zheng Y. A facile approach for fabrication of three-dimensional platinum-nanoporous gold film and its application for sensitive detection of microRNA-126 combining with catalytic hairpin assembly reaction. J Electroanal Chem. 2021;886: 115109.
Gong P, Levicky R. DNA surface hybridization regimes. Proc Natl Acad Sci. 2008;105:5301–6.
Zhang Y, Wang Y, Wang H, Jiang J-H, Shen G-L, Yu R-Q, Li J. Electrochemical DNA biosensor based on the proximity-dependent surface hybridization assay. Anal Chem. 2009;81:1982–7.
Li X, Dou B, Yuan R, Xiang Y. Mismatched catalytic hairpin assembly and ratiometric strategy for highly sensitive electrochemical detection of microRNA from tumor cells. Sens Actuat B Chem. 2019;286:191–7.
Huang PJ, Liu J. Signaling kinetics of DNA and aptamer biosensors revealing graphene oxide surface heterogeneity. J Anal Test. 2022;6(1):20–7.
SantaLucia J Jr, Hicks D. The thermodynamics of DNA structural motifs. Annu Rev Biophys Biomol Struct. 2004;33:415–40.
Knez K, Spasic D, Janssen KP, Lammertyn J. Emerging technologies for hybridization based single nucleotide polymorphism detection. Analyst. 2014;139:353–70.
Yan Y, Zhao D, Zhang Q, Chang YY, Liu M. A Thermophilic G-Quadruplex DNA/N- methylmesoporphyrin IX sensor for accurately detecting single nucleotide polymorphism. J Anal Test. 2022;6(1):53–9.
Hirajima S, Komatsu S, Ichikawa D, Takeshita H, Konishi H, Shiozaki A, Morimura R, Tsujiura M, Nagata H, Kawaguchi T. Clinical impact of circulating miR-18a in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer. 2013;108:1822–9.
Zouari M, Campuzano S, Pingarrón JM, Raouafi N. Femtomolar direct voltammetric determination of circulating miRNAs in sera of cancer patients using an enzymeless biosensor. Anal Chim Acta. 2020;1104:188–98.
Labib M, Khan N, Ghobadloo SM, Cheng J, Pezacki JP, Berezovski MV. Three-mode electrochemical sensing of ultralow microRNA levels. J Am Chem Soc. 2013;135:3027–38.
Acknowledgements
This work was supported by the Science and Technology Development Fund, Macau SAR (0040/2021/AGJ). The authors also thank the Department of Science and Technology of Guangdong Province for the support of Guangdong-Hong Kong-Macao Joint Laboratory of Respiratory Infectious Disease.
Author information
Authors and Affiliations
Contributions
FY: conceptualization, methodology, software, writing—original draft. YW: methodology, software. WZ: supervision, writing—review and editing. XB: conceptualization, methodology, supervision, writing—review and editing.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no known financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Yu, F., Wang, Y., Yu, C. et al. Sensitive and Specific Y-Shaped Ratio Biosensor for Detecting Serum miR-18a: Potential Early Scanning Tool for Non-Small Cell Lung Cancer. J. Anal. Test. 8, 237–244 (2024). https://doi.org/10.1007/s41664-023-00284-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41664-023-00284-w