Abstract
Untargeted metabolomics aims to comprehensively profile metabolites as many as possible in biological samples. Recently, ion mobility-mass spectrometry (IM-MS) has emerged as a powerful technology for untargeted metabolomics. The emerging role of IM-MS in untargeted metabolomics enables the separation of metabolite isomers and generation of multidimension data to support the identification of metabolites. In this review, we first introduced the basic principles of IM-MS instruments commonly used for untargeted metabolomics. Then, we demonstrated the application of IM-MS for metabolite separation and identification of both known and unknown metabolites. Finally, we discussed the future developments of IM-MS technology to improve untargeted metabolomics.


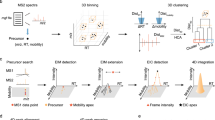
reproduced from Ref. 22 with permission from the American Chemical Society; c was reproduced from Ref. 47 with permission from the Royal Society of Chemistry)

reproduced from Ref. 50 with permission from the Royal Society of Chemistry; c was reproduced from Ref. 62 with permission from the American Chemical Society)
Similar content being viewed by others
Data Availability
Not applicable.
References
Nicholson JK, Lindon JC. Metabonomics. Nature. 2008;455(7216):1054–6.
Patti GJ, Yanes O, Siuzdak G. Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13(4):263–9. https://doi.org/10.1038/nrm3314.
Fiehn O. Metabolomics—the link between genotypes and phenotypes. In: Functional genomics. Springer, 2002; pp 155–171.
Kitano H. Systems biology: a brief overview. Science. 2002;295(5560):1662–4. https://doi.org/10.1126/science.1069492.
Cajka T, Fiehn O. Toward merging untargeted and targeted methods in mass spectrometry-based metabolomics and lipidomics. Anal Chem. 2016;88(1):524–45.
Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol. 2004;5(9):763–9.
Zhu ZJ, Schultz AW, Wang J, Johnson CH, Yannone SM, Patti GJ, Siuzdak G. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat Protoc. 2013;8(3):451–60.
Vinaixa M, Schymanski EL, Neumann S, Navarro M, Salek RM, Yanes O. Mass spectral databases for LC/MS- and GC/MS-based metabolomics: state of the field and future prospects. TrAC, Trends Anal Chem. 2016;78:23–35.
Nichols CM, Dodds JN, Rose BS, Picache JA, Morris CB, Codreanu SG, May JC, Sherrod SD, McLean JA. Untargeted molecular discovery in primary metabolism: collision cross section as a molecular descriptor in ion mobility-mass spectrometry. Anal Chem. 2018;90(24):14484–92. https://doi.org/10.1021/acs.analchem.8b04322.
Lapthorn C, Pullen F, Chowdhry BZ. Ion mobility spectrometry-mass spectrometry (IMS-MS) of small molecules: separating and assigning structures to ions. Mass Spectrom Rev. 2013;32(1):43–71.
May JC, Gant-Branum RL, McLean JA. Targeting the untargeted in molecular phenomics with structurally-selective ion mobility-mass spectrometry. Curr Opin Biotechnol. 2016;39:192–7.
Paglia G, Astarita G. Metabolomics and lipidomics using traveling-wave ion mobility mass spectrometry. Nat Protoc. 2017;12(4):797–813. https://doi.org/10.1038/nprot.2017.013.
Lanucara F, Holman SW, Gray CJ, Eyers CE. The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nat Chem. 2014;6(4):281.
May JC, McLean JA. Ion mobility-mass spectrometry: time-dispersive instrumentation. Anal Chem. 2015;87(3):1422–36.
Zheng X, Wojcik R, Zhang X, Ibrahim YM, Burnum-Johnson KE, Orton DJ, Monroe ME, Moore RJ, Smith RD, Baker ES. Coupling front-end separations, ion mobility spectrometry, and mass spectrometry for enhanced multidimensional biological and environmental analyses. Ann Rev Anal Chem. 2017;10:71–92.
May JC, Morris CB, McLean JA. Ion mobility collision cross section compendium. Anal Chem. 2016;89(2):1032–44. https://doi.org/10.1021/acs.analchem.6b04905.
Paglia G, Williams JP, Menikarachchi L, Thompson JW, Tyldesley-Worster R, Halldórsson S, Rolfsson O, Moseley A, Grant D, Langridge J, Palsson BQ, Astarita G. Ion mobility derived collision cross sections to support metabolomics applications. Anal Chem. 2014;86(8):3985–93. https://doi.org/10.1021/ac500405x.
Zhang X, Kew K, Reisdorph R, Sartain M, Powell R, Armstrong M, Quinn K, Cruickshank-Quinn C, Walmsley S, Bokatzian S, Darland E, Rain M, Imatani K, Reisdorph N. Performance of a high-pressure liquid chromatography–ion mobility-mass spectrometry system for metabolic profiling. Anal Chem. 2017;89(12):6384–91. https://doi.org/10.1021/acs.analchem.6b04628.
Zhou Z, Tu J, Zhu ZJ. Advancing the large-scale CCS database for metabolomics and lipidomics at the machine-learning era. Curr Opin Chem Biol. 2018;42:34–41.
Kolakowski BM, Mester Z. Review of applications of high-field asymmetric waveform ion mobility spectrometry (FAIMS) and differential mobility spectrometry (DMS). Analyst. 2007;132(9):842–64.
Zhang JD, Mohibul Kabir KM, Lee HE, Donald WA. Chiral recognition of amino acid enantiomers using high-definition differential ion mobility mass spectrometry. Int J Mass Spectrom. 2018;428:1–7. https://doi.org/10.1016/j.ijms.2018.02.003.
May JC, Goodwin CR, Lareau NM, Leaptrot KL, Morris CB, Kurulugama RT, Mordehai A, Klein C, Barry W, Darland E, Overney G, Imatani K, Stafford GC, Fjeldsted JC, McLean JA. Conformational ordering of biomolecules in the gas phase: nitrogen collision cross sections measured on a prototype high resolution drift tube ion mobility-mass spectrometer. Anal Chem. 2014;86(4):2107–16. https://doi.org/10.1021/ac4038448.
Schroeder M, Meyer SW, Heyman HM, Barsch A, Sumner LW. Generation of a collision cross section library for multi-dimensional plant metabolomics using UHPLC-trapped ion mobility-MS/MS. Metabolites. 2019;10(1):13. https://doi.org/10.3390/metabo10010013.
Dodds JN, Baker ES. Ion mobility spectrometry: fundamental concepts, instrumentation, applications, and the road ahead. J Am Soc Mass Spectrom. 2019;30(11):2185–95.
Gabelica V, Shvartsburg AA, Afonso C, Barran P, Benesch JLP, Bleiholder C, Bowers MT, Bilbao A, Bush MF, Campbell JL, Campuzano IDG, Causon T, Clowers BH, Creaser CS, Pauw ED, Far J, Fernandez-Lima F, Fjeldsted JC, Giles K, Groessl M, Hogan CJ Jr, Hann S, Kim HI, Kurulugama RT, May JC, McLean JA, Pagel K, Richardson K, Ridgeway ME, Rosu F, Sobott F, Thalassinos K, Valentine SJ, Wyttenbach T. Recommendations for reporting ion mobility mass spectrometry measurements. Mass Spectrom Rev. 2019;38(3):291–32020. https://doi.org/10.1002/mas.21585.
Kurulugama RT, Darland E, Kuhlmann F, Stafford G, Fjeldsted J. Evaluation of drift gas selection in complex sample analyses using a high performance drift tube ion mobility-QTOF mass spectrometer. Analyst. 2015;140(20):6834–44.
Stow SM, Causon TJ, Zheng X, Kurulugama RT, Mairinger T, May JC, Rennie EE, Baker ES, Smith RD, McLean JA, Hann S, Fjeldsted JC. An interlaboratory evaluation of drift tube ion mobility–mass spectrometry collision cross section measurements. Anal Chem. 2017;89(17):9048–55.
Hines KM, Ross DH, Davidson KL, Bush MF, Xu L. Large-scale structural characterization of drug and drug-like compounds by high-throughput ion mobility-mass spectrometry. Anal Chem. 2017;89(17):9023–30. https://doi.org/10.1021/acs.analchem.7b01709.
Zheng X, Aly NA, Zhou Y, Dupuis KT, Bilbao A, Paurus VL, Orton DJ, Wilson R, Payne SH, Smith RD, Baker ES. A structural examination and collision cross section database for over 500 metabolites and xenobiotics using drift tube ion mobility spectrometry. Chem Sci. 2017;8(11):7724–36. https://doi.org/10.1039/c7sc03464d.
Zhou Z, Shen X, Tu J, Zhu ZJ. Large-scale prediction of collision cross-section values for metabolites in ion mobility-mass spectrometry. Anal Chem. 2016;88(22):11084–91. https://doi.org/10.1021/acs.analchem.6b03091.
Zhou Z, Tu J, Xiong X, Shen X, Zhu ZJ. LipidCCS: prediction of collision cross-section values for lipids with high precision to support ion mobility–mass spectrometry-based lipidomics. Anal Chem. 2017;89(17):9559–666. https://doi.org/10.1021/acs.analchem.7b02625.
Bush MF, Campuzano ID, Robinson CV. Ion mobility mass spectrometry of peptide ions: effects of drift gas and calibration strategies. Anal Chem. 2012;84(16):7124–30.
Hines KM, May JC, McLean JA, Xu L. Evaluation of collision cross section calibrants for structural analysis of lipids by traveling wave ion mobility-mass spectrometry. Anal Chem. 2016;88(14):7329–36.
Giles K, Ujma J, Wildgoose J, Pringle S, Richardson K, Langridge D, Green M. A cyclic ion mobility-mass spectrometry system. Anal Chem. 2019;91(13):8564–73. https://doi.org/10.1021/acs.analchem.9b01838.
Tolmachev AV, Webb IK, Ibrahim YM, Garimella SV, Zhang X, Anderson GA, Smith RD. Characterization of ion dynamics in structures for lossless ion manipulations. Anal Chem. 2014;86(18):9162–8.
Vasilopoulou CG, Sulek K, Brunner AD, Meitei NS, Schweiger-Hufnagel U, Meyer SW, Barsch A, Mann M, Meier F. Trapped ion mobility spectrometry and PASEF enable in-depth lipidomics from minimal sample amounts. Nat Commun. 2020;11(1):1–11.
Jeanne Dit Fouque K, Ramirez CE, Lewis RL, Koelmel JP, Garrett TJ, Yost RA, Fernandez-Lima F. Effective liquid chromatography-trapped ion mobility spectrometry-mass spectrometry separation of isomeric lipid species. Anal Chem. 2019;91(8):5021–7. https://doi.org/10.1021/acs.analchem.8b04979.
Hernandez-Mesa M, Le Bizec B, Monteau F, Garcia-Campana AM, Dervilly-Pinel G. Collision cross section (CCS) database: an additional measure to characterize steroids. Anal Chem. 2018;90(7):4616–25. https://doi.org/10.1021/acs.analchem.7b05117.
Dodds JN, May JC, McLean JA. Investigation of the complete suite of the leucine and isoleucine isomers: toward prediction of ion mobility separation capabilities. Anal Chem. 2016;89(1):952–9. https://doi.org/10.1021/acs.analchem.6b04171.
Leaptrot KL, May JC, Dodds JN, McLean JA. Ion mobility conformational lipid atlas for high confidence lipidomics. Nat Commun. 2019;10(1):985. https://doi.org/10.1038/s41467-019-08897-5.
Ross DH, Seguin RP, Xu L. Characterization of the impact of drug metabolism on the gas-phase structures of drugs using ion mobility-mass spectrometry. Anal Chem. 2019;91(22):14498–50707. https://doi.org/10.1021/acs.analchem.9b03292.
Zheng X, Dupuis KT, Aly NA, Zhou Y, Smith FB, Tang K, Smith RD, Baker ES. Utilizing ion mobility spectrometry and mass spectrometry for the analysis of polycyclic aromatic hydrocarbons, polychlorinated biphenyls, polybrominated diphenyl ethers and their metabolites. Anal Chim Acta. 2018;1037:265–73.
Wojcik R, Webb I, Deng L, Garimella S, Prost S, Ibrahim Y, Baker ES, Smith RD. Lipid and glycolipid isomer analyses using ultra-high resolution ion mobility spectrometry separations. Int J Mol Sci. 2017;18(1):183. https://doi.org/10.3390/ijms18010183.
Poad BLJ, Zheng X, Mitchell TW, Smith RD, Baker ES, Blanksby SJ. Online ozonolysis combined with ion mobility-mass spectrometry provides a new platform for lipid isomer analyses. Anal Chem. 2018;90(2):1292–300. https://doi.org/10.1021/acs.analchem.7b04091.
Hofmann J, Hahm HS, Seeberger PH, Pagel K. Identification of carbohydrate anomers using ion mobility–mass spectrometry. Nature. 2015;526(7572):241–4. https://doi.org/10.1038/nature15388.
Wu Q, Wang JY, Han DQ, Yao ZP. Recent advances in differentiation of isomers by ion mobility mass spectrometry. TrAC Trends in Anal Chem. 2020;124:115801. https://doi.org/10.1016/j.trac.2019.115801.
Domalain V, Hubert-Roux M, Tognetti V, Joubert L, Lange CM, Rouden J, Afonso C. Enantiomeric differentiation of aromatic amino acids using traveling wave ion mobility-mass spectrometry. Chem Sci. 2014;5(8):3234–9. https://doi.org/10.1039/c4sc00443d.
Yu X, Yao ZP. Chiral differentiation of amino acids through binuclear copper bound tetramers by ion mobility mass spectrometry. Anal Chim Acta. 2017;981:62–70. https://doi.org/10.1016/j.aca.2017.05.026.
McCullagh M, Douce D, Van Hoeck E, Goscinny S. Exploring the complexity of steviol glycosides analysis using ion mobility mass spectrometry. Anal Chem. 2018;90(7):4585–95. https://doi.org/10.1021/acs.analchem.7b05002.
Picache JA, Rose BS, Balinski A, Leaptrot KL, Sherrod SD, May JC, McLean JA. Collision cross section compendium to annotate and predict multi-omic compound identities. Chem Sci. 2019;10(4):983–93. https://doi.org/10.1039/c8sc04396e.
Plante PL, Francovic-Fontaine E, May JC, McLean JA, Baker ES, Laviolette F, et al. Predicting ion mobility collision cross-sections using a deep neural network: DeepCCS. Anal Chem. 2019;91(8):5191–9. https://doi.org/10.1021/acs.analchem.8b05821.
Bijlsma L, Bade R, Celma A, Mullin L, Cleland G, Stead S, Hernandez F, Sancho JV. Prediction of collision cross-section values for small molecules: application to pesticide residue analysis. Anal Chem. 2017;89(12):6583–9. https://doi.org/10.1021/acs.analchem.7b00741.
Colby SM, Nunez JR, Hodas NO, Corley CD, Renslow RR. Deep learning to generate in silico chemical property libraries and candidate molecules for small molecule identification in complex samples. Anal Chem. 2020;92(2):1720–9. https://doi.org/10.1021/acs.analchem.9b02348.
Colby SM, Thomas DG, Nuñez JR, Baxter DJ, Glaesemann KR, Brown JM, Pirrung MA, Govind N, Teeguarden JG, Metz TO, Renslow RS. ISiCLE: a quantum chemistry pipeline for establishing in silico collision cross section libraries. Anal Chem. 2019;91(7):4346–56. https://doi.org/10.1021/acs.analchem.8b04567.
Mesleh M, Hunter J, Shvartsburg A, Schatz GC, Jarrold M. Structural information from ion mobility measurements: effects of the long-range potential. J Phys Chem. 1996;100(40):16082–6.
Zhou Z, Luo M, Chen X, Yin Y, Xiong X, Zhu ZJ. AllCCS Web server. https://allccs.zhulab.cn/. Accessed 25 April 2020.
Stephan S, Hippler J, Köhler T, Deeb AA, Schmidt TC, Schmitz OJ. Contaminant screening of wastewater with HPLC-IM-qTOF-MS and LC+LC-IM-qTOF-MS using a CCS database. Anal Bioanal Chem. 2016;408(24):6545–55. https://doi.org/10.1007/s00216-016-9820-5.
Regueiro J, Negreira N, Berntssen MH. Ion-mobility-derived collision cross section as an additional identification point for multiresidue screening of pesticides in fish feed. Anal Chem. 2016;88(22):11169–77. https://doi.org/10.1021/acs.analchem.6b03381.
Zhou Z, Shen X, Chen X, Tu J, Xiong X, Zhu ZJ. LipidIMMS analyzer: integrating multi-dimensional information to support lipid identification in ion mobility—mass spectrometry based lipidomics. Bioinformatics. 2019;35(4):698–700.
Tsugawa H, Ikeda K, Takahashi M, Satoh A, Mori Y, Uchino H, Okahashi N, Yamada Y, Tada I, Bonini P, Higashi Y, Okazaki Y, Zhou ZW, Zhu ZJ, Koelmel J, Cajka T, Fiehn O, Saito K, Arita M, Arita M. MS-DIAL 4: accelerating lipidomics using an MS/MS, CCS, and retention time atlas. bioRxiv. 2020:2020.02.11.944900. doi:10.1101/2020.02.11.944900.
Goodwin CR, Fenn LS, Derewacz DK, Bachmann BO, McLean JA. Structural mass spectrometry: rapid methods for separation and analysis of peptide natural products. J Nat Prod. 2012;75(1):48–53. https://doi.org/10.1021/np200457r.
Bijlsma L, Berntssen MHG, Merel S. A refined nontarget workflow for the investigation of metabolites through the prioritization by in silico prediction tools. Anal Chem. 2019;91(9):6321–8. https://doi.org/10.1021/acs.analchem.9b01218.
Trim PJ, Henson CM, Avery JL, McEwen A, Snel MF, Claude E, Marshell PS, West A, Princivalle AP, Clench MR. Matrix-assisted laser desorption/ionization-ion mobility separation-mass spectrometry imaging of vinblastine in whole body tissue sections. Anal Chem. 2008;80(22):8628–34.
Škrášková K, Claude E, Jones EA, Towers M, Ellis SR, Heeren RM. Enhanced capabilities for imaging gangliosides in murine brain with matrix-assisted laser desorption/ionization and desorption electrospray ionization mass spectrometry coupled to ion mobility separation. Methods. 2016;104:69–78.
Li H, Smith BK, Márk L, Nemes P, Nazarian J, Vertes A. Ambient molecular imaging by laser ablation electrospray ionization mass spectrometry with ion mobility separation. Int J Mass Spectrom. 2015;377:681–9.
Causon TJ, Si-Hung L, Newton K, Kurulugama RT, Fjeldsted J, Hann S. Fundamental study of ion trapping and multiplexing using drift tube-ion mobility time-of-flight mass spectrometry for non-targeted metabolomics. Anal Bioanal Chem. 2019;411(24):6265–74. https://doi.org/10.1007/s00216-019-02021-8.
Zang X, Monge ME, Gaul DA, Fernandez FM. Flow injection-traveling-wave ion mobility-mass spectrometry for prostate-cancer metabolomics. Anal Chem. 2018;90(22):13767–74. https://doi.org/10.1021/acs.analchem.8b04259.
Zhang X, Romm M, Zheng X, Zink EM, Kim YM, Burnum-Johnson KE, Orton DJ, Apffel A, Ibrahim YM, Monroe ME, Moore RJ, Smith JN, Ma J, Renslow RS, Thomas DG, Blackwell AE, Swinford G, Sausen J, Kurulugama RT, Eno N, Darland E, Stafford G, Fjeldsted J, Metz TO, Teeguarden JG, Smith RD, Baker ES. SPE-IMS-MS: an automated platform for sub-sixty second surveillance of endogenous metabolites and xenobiotics in biofluids. Clin Mass Spectrom. 2016;2:1–10. https://doi.org/10.1016/j.clinms.2016.11.002.
Zhang L, Foreman DP, Grant PA, Shrestha B, Moody SA, Villiers F, Kwak JM, Vertes A. In situ metabolic analysis of single plant cells by capillary microsampling and electrospray ionization mass spectrometry with ion mobility separation. Analyst. 2014;139(20):5079–85. https://doi.org/10.1039/c4an01018c.
Acknowledgements
The work was supported by National Natural Science Foundation of China (Grant No. 31971356), and Shanghai Municipal Science and Technology Major Project (Grant No. 2019SHZDZX02).
Funding
The work was supported by National Natural Science Foundation of China (Grant No. 31971356), and Shanghai Municipal Science and Technology Major Project (Grant No. 2019SHZDZX02).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
About this article
Cite this article
Luo, MD., Zhou, ZW. & Zhu, ZJ. The Application of Ion Mobility-Mass Spectrometry in Untargeted Metabolomics: from Separation to Identification. J. Anal. Test. 4, 163–174 (2020). https://doi.org/10.1007/s41664-020-00133-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41664-020-00133-0