Abstract
Plant diseases cause significant agricultural losses, demanding accurate detection methods. Traditional approaches relying on expert knowledge may be biased, but advancements in computing, particularly deep learning, offer non-experts effective tools. This study focuses on fine-tuning cutting-edge pre-trained CNN and vision transformer models to classify grape leaves and diagnose grape leaf diseases through digital images. Our research examined a PlantVillage dataset, which comprises 4062 leaf images distributed across four categories. Additionally, we utilized the Grapevine dataset, consisting of 500 leaf images. This dataset is organized into five distinct groups, with each group containing 100 images corresponding to one of the five grape types. The PlantVillage dataset focuses on four classes related to grape diseases, namely Black Rot, Leaf Blight, Healthy, and Esca leaves. On the other hand, the Grapevine dataset includes five classes for leaf recognition, specifically Ak, Alaidris, Buzgulu, Dimnit, and Nazli. In experiments with 14 CNN and 17 vision transformer models, deep learning demonstrated high accuracy in distinguishing grape diseases and recognizing leaves. Notably, four models achieved 100% accuracy on PlantVillage and Grapevine datasets, with Swinv2-Base standing out. This approach holds promise for enhancing crop productivity through early disease detection and providing insights into grape variety characterization in agriculture.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
Introduction
Agriculture is the production of plant and animal products using soil and seeds and their evaluation at various stages. Agriculture, in a broader sense, encompasses activities undertaken in a specific biological and socio-economic environment to obtain the necessary animal and plant products for human nutrition (Sandhu 2021). Today, agriculture is one of the most important strategic sectors in the world. In the coming years, it is expected that the world population will increase significantly in developing countries, dietary habits will change globally, new technologies will emerge, and while developed countries increasingly turn to organic foods, there may be a struggle with genetically modified foods in developing countries (Hekimoğlu and Altındeğer 2006).
One of the most consumed fruits in the world, grapes are also the major component used to make wine. Grape output and quality therefore have a high economic value (Peng et al. 2021). However, grape leaves are prone to a number of illnesses brought on by the environment, the weather, and most commonly by fungi, viruses, and bacteria (Armijo et al. 2016). Ineffective control of grape leaf diseases causes the disease to spread throughout the plant, which lowers grape quality and output. Classic phytopathology techniques were initially used to identify the grape leaf disease (Ji et al. 2020); however, manual identification techniques are labor- and time-intensive (Xiao et al. 2023). The reliability of manual identification methods decreases as more acreage is used for grape production. For the development of grape production in future, automatic recognition of grape leaf diseases is crucial (Singh and Misra 2017). For these reasons, this study focuses on diagnosing grape diseases and grape types from grape leaves.
In today's agriculture, the goal is to produce the highest output possible with the least number of resources and effort. So, deep learning, machine learning, and image processing methods of the present day are employed. To increase earnings from agricultural products, precise quality control is required (Ghoury et al. 2019). Today's modern agriculture aims to produce the highest yield possible with the least amount of energy and effort. As a result, cutting-edge image processing, machine learning, and deep learning methods are employed. To increase revenues, agricultural products need precise quality control (Muthukannan and Latha 2018; Murakami et al. 2005; Paulus et al. 1997), and diagnosis and identification of plant pests (Abbasgholipour et al. 2011; Boissard et al. 2008; Shariff et al. 2006). Around the world, plant diseases constitute a key factor in crop losses (Aggarwal and Jaidka 2022). The key to sustainable agriculture is plant disease identification, which has grown in importance in the agricultural industry (Martinelli et al. 2015; Pydipati et al. 2006). To guarantee the quality of the harvest, it is essential to identify and diagnose plant diseases quickly and accurately, as well as to put the required controls in place (Zhu et al. 2020). Considering these reasons, the automatic detection of grape and grape leaf diseases using an automated system is important. Identifying plant diseases using images is a significant challenge. In recent years, several plant diseases and crops have seen encouraging outcomes from the classification of plant diseases using various machine learning methods (Ghoury et al. 2019).
Deep learning is a machine learning method that can make highly accurate predictions by working with large datasets, especially consisting of artificial neural networks (Aggarwal 2020; Coşkun et al. 2023). Artificial neural networks are designed based on the principles of the human brain, and deep learning is achieved by making these artificial neural networks multi-layered. This allows for solving more complex and abstract problems. Deep learning techniques have shown remarkable results, particularly in areas such as image and speech recognition. Deep learning is an important tool for increasing agricultural productivity. Farmers need to monitor many factors such as plant growth, productivity, disease, and pest control (Veziroglu et al. 2023). Deep learning can make more accurate predictions by analyzing the complex relationships among these factors. For example, plant growth rate can be predicted using field data and weather data, or disease symptoms can be detected. These applications help improve agricultural productivity.
The increasing use of artificial intelligence applications in agriculture provides solutions to plant identification and classification problems (Eli-Chukwu 2019). Recent years have seen the successful application of plant diagnosis systems in fields including yield prediction, disease identification, and species estimation (Patrício and Rieder 2018; Manavalan 2020). Leaves are the most suitable option for plant identification as they contain species-specific features, are abundantly available, and present throughout a significant portion of the year (Mishra et al. 2012). Leaf image analysis and machine learning techniques are widely employed in plant classification (Koklu and Ozkan 2020; Kaya and Saritas 2019). However, existing methods have limitations and there is a need for more advanced approaches that provide deeper insights. This is where deep learning approaches come into the picture. Deep learning is a more recent method in computer vision that can extract more precise data. It can use leaf features that are automatically extracted to classify objects. Numerous deep learning-based leaf categorization techniques have been proposed in recent years (Beikmohammadi and Faez 2018; Tavakoli et al. 2021).
Deep learning also enables more efficient use of resources. Resources like water and fertilizer yield the best results when applied in the correct amounts and at the right time. Deep learning algorithms can analyze plant requirements and soil characteristics to develop optimal irrigation and fertilization strategies (Kiliçarslan and Pacal 2023). This reduces water and fertilizer usage in agricultural production and allows for sustainable farming practices. Deep learning also plays an important role in the control of agricultural diseases and pests. Early detection of diseases and pests is crucial for swift intervention and implementation of control measures. Deep learning algorithms can quickly diagnose plant diseases by analyzing symptoms and images of pests. This enables farmers to intervene in a timely manner and minimize potential losses. As a sequence, the use of deep learning in agriculture provides several advantages to farmers. Increased productivity, more efficient resource utilization, and enhanced capabilities in disease/pest control are important tools for improving agricultural production and implementing sustainable farming practices.
The primary objective of our study is to employ deep learning methods for the classification of grape diseases and grape leaves. By leveraging advanced techniques in deep learning, we aim to develop a robust and accurate system capable of distinguishing various types of grape diseases as well as classifying different grape leaf categories. This research seeks to contribute to the field of agricultural technology by providing an effective tool for early detection and classification of issues affecting grape crops. Through the utilization of deep learning algorithms, we endeavor to enhance the efficiency and precision of grape disease and leaf classification, ultimately aiding vineyard management practices. In summary, our main objective is to harness the power of deep learning to advance the capabilities of grape disease and leaf classification systems for the benefit of the agricultural community.
Related works
The identification and classification of plant diseases are crucial aspects of the agriculture industry, with far-reaching impacts on crop yield, quality, and overall agricultural productivity. However, the field has witnessed significant advancements in image processing, machine learning, and artificial neural network technologies, leading to substantial progress in plant disease recognition and classification. Notably, the application of CNN (convolutional neural network) and vision transformer algorithms has emerged as a particularly successful approach in tackling these challenges by automatically extracting features for image recognition tasks. Several studies in this domain have contributed valuable insights and breakthroughs, which are summarized as follows.
In their study, Tang et al. (2020) proposed new model architectures despite the use of traditional CNN for the automatic diagnosis of plant diseases. Five different CNN-based models were tested, and ShuffleNet was found to be the most suitable. Compared to the baseline model, the suggested model showed improved discriminative ability and classification accuracy. The advantages of the model included the use of squeeze-and-excitation blocks to achieve high-quality spatial encoding and reduce computational costs. The model achieved real-time and highly accurate classification of diseased grape leaves (99.14%). Future research will focus on combining the suggested algorithm with an IoT platform to improve the efficiency of the automated process.
In the article by Jin et al. (2022), it was noted that the use of the GAN method was effective in increasing the dataset of grape leaf disease images, but the generated images lacked clarity. To address these issues, a new image enhancement method called GrapeGAN was proposed. GrapeGAN utilized a generator and a discriminator in a U-Net-like structure designed to preserve fine-grained features and enhance the structural integrity of the generated images. Experimental results demonstrated that the images of grape leaf disease produced by GrapeGAN were of higher quality compared to other methods and achieved successful recognition tasks. Therefore, the GrapeGAN method exhibits good applicability in improving grape leaf disease images and aims to overcome the limitations of traditional GAN generation methods.
Ji et al. (2020) emphasized that grape diseases cause significant losses, making it an urgent requirement to develop an automated diagnostic method. Given the impressive achievements of deep learning techniques in computer vision problems, the article proposes a unified CNN. Black rot, esca, and leaf blight leaf spot are a few common grape diseases that this architecture is intended to separate from healthy leaves. The UnitedModel, which was evaluated on the PlantVillage dataset, is anticipated to improve representational capability as a result of the merging of numerous CNNs. UnitedModel achieved a higher accuracy rate compared to other CNN models and, consequently, can serve as a decision-support tool to assist farmers in identifying grape diseases.
For the automatic detection of grape downy mildew and grape powdery mildew diseases and the prompt delivery of pertinent information, Li et al. (2012) used image recognition technology. The K-means clustering technique was used to unsupervisedly cluster disease images, and fifty various features were then retrieved from the disease images. A SVM classifier was designed based on 31 effectively selected features for disease classification, achieving training recognition rates of 100% for both grape diseases, while the test recognition rates were 90% and 93.33%, respectively. The recognition results from SVMs with various kernels demonstrated that the linear kernel SVM was best suited for identifying diseases from images. This study provides a basis and point of reference for the creation of an automatic diagnosis system for plant diseases, as well as an efficient method for the quick and accurate identification and diagnosis of plant illnesses.
In the study by Liu et al. (2020), a new model called Leaf GAN is proposed for the identification of grape leaf diseases using deep learning. The model generates images of four different grape leaf diseases using generative adversarial networks (GANs). Using a dense connectivity technique and instance normalization, the model's discriminator can tell real disease images from phony ones. The Leaf GAN model ultimately produces 8,124 disease images from a total of 4062 images of grape leaf disease. The deep regret gradient penalty method stabilizes the model's training. According to experimental findings, the Leaf GAN model generates an adequate number of images of grape leaf disease and offers a workable method for data augmentation. Experiments using an extended dataset and eight classification models demonstrate that CNN-based identification achieves higher accuracies. The suggested data augmentation method provides a fresh way to deal with overfitting problems in disease detection and significantly raises the accuracy of identification.
An algorithm for automatic crop detection was put out by Hamuda et al. in 2017. Under various weather circumstances and in natural light, this system was utilized to identify cauliflowers in video streams, and the identification results were contrasted with actual data collected through manual labeling. The algorithm achieved 99.04% accuracy and 98.91% sensitivity. A technique based on support vector machines was put out by Akbarzadeh et al. (2018) for classifying plants. The experimental findings demonstrated that the suggested algorithm successfully categorized plants with 97% accuracy. A technique for identifying cucumber powdery mildew based on visual spectra was proposed by Zhang and Wang (2016). After being identified and categorized, the spectral features, and the visible light band between 450 and 780 nm was chosen as the research range. Then, a classification model was created using the SVM algorithm, and the kernel function was optimized using a radial basis function. According to the experimental findings, this model's accuracy rates were 100% and 96.25%, respectively, for a total accuracy of 98.13%.
In the study by Waghmare et al. (2016), a method for the analysis of leaf texture and pattern recognition was proposed for the identification of grape diseases. The system receives a single plant leaf as input, and segmentation is carried out after background removal. A high-pass filter is used to examine the segmented leaf image and find the sick area. Finally, a multi-class SVM is given the extracted texture pattern. An intelligent technique for the identification of grape fanleaf virus was put forth by Mohammadpoor et al. (2020). The fuzzy C-mean approach is used to highlight each leaf's sick areas, which are subsequently identified using SVM.
CNN eliminates the need for image preprocessing and feature extraction steps, which are considered unnecessary by machine learning algorithms, as it automatically identifies and extracts distinctive features for image recognition (Kulin et al. 2018; Zhang et al. 2018).
Numerous studies have demonstrated the successful use of CNN algorithms for plant disease recognition. For example, in the study by Fuentes et al. (2017), a CNN-based approach was proposed for detecting diseases in tomato plants, achieving a high accuracy rate of 98.4%. In the study by Ferentinos (2018), analysis of simple leaf images of healthy and diseased plants for the detection and diagnosis of plant diseases was performed using CNN models. The models were trained with a database containing 87,848 images and achieved the best performance with a success rate of 99.53%. This high success rate indicates that the model can be used as a useful advisory tool or an early warning system and has the potential to be extended into an integrated plant disease recognition system under real cultivation conditions.
Material and methods
Datasets
The importance of the dataset in achieving the success of deep learning architectures is undeniable. Compared to classical machine learning approaches, the large-scale dataset and automatic feature extraction in deep learning architectures represent significant differences. This is because deep learning architectures inherently have a hunger for data-driven features (Pacal 2024a).
PlantVillage dataset
In this study, the largest publicly available dataset containing numerous plant diseases, namely the PlantVillage dataset, was used to effectively detect diseases in grape leaves. The PlantVillage dataset includes 54,303 leaf images categorized into 38 species and disease groups, including both healthy and unhealthy samples (Hughes and Salathé, 2015). In our research, we focused on the grape-related categories in this dataset to examine the diagnosis of diseases in grape leaves. The PlantVillage dataset has four categories related to grape leaf diseases. In this study, we analyzed these four categories, which contain 4062 leaf images.
Figure 1 provides the features of healthy and diseased leaf images in the PlantVillage dataset for each class. Table 1 provides a detailed overview of the characteristics of each grape leaf disease category in the PlantVillage dataset.
As seen in Table 1, the features of the four different grape leaf classes in the PlantVillage dataset used in the study and the data partitioning method are described. The leaf images, each in a separate folder, are located in a single folder without a distinction between validation and test data. To measure the generalization ability of the models and make the study more objective, the data is divided into 70% training, 15% validation, and 15% test for each class. While most studies in the literature only use training and validation data, this study creates a more ideal dataset for deep learning architectures by using all three subsets of data.
Grapevine dataset
In this study, a publicly available dataset called the Grapevine dataset is used to more effectively detect the type of grapes from grape leaves. The Grapevine dataset consists of 500 leaf images, with 100 images for each of the five grape types in separate groups (Koklu et al. 2022). In our research, we used this dataset to examine the use of grape leaves for type diagnosis. Figure 2 provides randomly selected samples from Grapevine dataset.
The grape types in the Grapevine dataset are 'ak', 'ala idris', 'buzgulu', 'dimnit', and 'nazlı'. In this study, we analyzed these five categories. Table 2 provides detailed information on the features of each grape class in the Grapevine dataset.
Deep learning architectures
Machine learning methods have achieved great accomplishments for the progress and modernization of society. They are widely used in various applications ranging from finding search queries on the web to filtering social media content and recommendations on e-commerce websites. Moreover, with advancing technology, they have become an active part of our daily lives through smart devices. Machine learning also includes applications such as recognizing objects in an image, converting spoken words into written text, and matching specific news or social media posts to users' interests (LeCun et al. 2015).
These applications utilize deep learning techniques, which are a branch of machine learning methods used in "deep" network architectures. Deep learning enables the learning of data using computational models and algorithms consisting of one or more layers. It can detect complex structures in large datasets using the backpropagation algorithm. These methods have propelled technologies developed in different fields, such as speech recognition, image recognition, and object detection, to the highest levels. Deep convolutional networks play a significant role in tasks like image, video, speech, and audio processing, while recurrent networks enable the discovery of sequential data like text and speech (LeCun et al. 2015).
Although the concept of deep learning emerged in 2006 (Hinton 2006), it gained widespread popularity, particularly through the ImageNet competition. The ImageNet competition provided a platform for showcasing algorithms for image recognition, where deep learning architectures stood out. Deep learning architectures have achieved remarkable results in the field of object recognition. Additionally, they are used in pattern recognition, detection, classification, future prediction, drug production, dictionary creation, signal processing, and medical and financial domains, as well as in defense industries. Studies have shown that deep learning structures yield significantly better results compared to other known methods (Pacal 2024b).
Convolutional neural networks
In the field of deep learning, the convolutional neural network (CNN) is a key model that is utilized in many applications. Especially used in image processing and recognition domains, CNN has a specialized structure and operates by employing convolutional, pooling, and fully connected layers on input data. The convolutional layers utilize filters to identify and extract features from the data as output. The pooling layers reduce the output size while scaling the features, and the fully connected layers are used for classification or prediction tasks.
Convolutional layer consists of a collection of trainable filters designed for conducting feature extraction. Assuming X represents the input data with k filters present in the convolutional layers, the output of the convolutional layer can be calculated in the following manner (Eq. 1). \({w}_{j}\) and \({b}_{j}\) represent the weight and bias, respectively, and f denotes an activation function. The symbol * signifies the convolution operation.
Pooling layer is used to reduce the dimensions of the obtained feature data and network parameters. Currently, the most used methods for this purpose are max pooling and average pooling (Eq. 2). Assuming S is a p × p window size, the average pooling operation can be defined as follows, where \({x}_{ij}\) represents the activation value at (i, j), and N is the total number of elements in S.
Fully connected layer: Following the last pooling layer, the fully connected layer is employed to transform the feature maps into a 1-D feature vector. This transformation can be represented as Y and z, denoting the output vector and input features (Eq. 3). In contrast, w and b represent the weight and bias of the fully connected layer, respectively.
The training of CNN is performed by minimizing the error function through the optimization of parameters, resulting in improved performance. Consequently, CNN is a powerful artificial neural network model used to achieve high accuracy rates in image processing and recognition domains. The typical algorithm for CNN is presented in Algorithm 1.
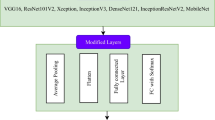
CNN consists of a convolutional layer that defines features using filters on input data, an activation function that non-linearly transforms the convolutional outputs, a pooling layer that reduces the outputs while preserving features, and fully connected layers. Following the convolution, activation, and pooling layers, fully connected layers are used for classification or prediction (Fig. 3). The training of CNN is carried out by minimizing the error function between the predicted values and the actual values.
Vision transformer
A vision transformer is a deep learning model specifically designed for computer vision tasks, leveraging self-attention mechanisms to capture long-range dependencies in image data. Unlike traditional CNNs, the vision transformer processes input images through a transformer architecture, demonstrating its effectiveness in image recognition and object detection. The transformer architecture was created for natural language processing (NLP) tasks, and ViT is based on it (Pacal and Kılıcarslan 2023). The key component of ViT is the self-attention mechanism, which models the interaction between different parts of the input image and focuses on important features. The input image is divided into patches of a certain size, and these patches interact with each other through attention heads and fully connected layers within transformer blocks. As a result, the output layer produces results for classification or another task (Fig. 4).
One of the main advantages of vision transformer is its ability to handle images of different sizes and effectively process data. However, it can be computationally expensive and demand a lot of memory and processing power.
In the ViT architecture, the input image is divided into fixed-size patches, and each patch transforms a continuous vector using a process called linear embedding. Additionally, positional embeddings are introduced to the patch representations to preserve positional information. Following this, a series of consecutive patches (Eq. 4) are input into the transformer encoder. The encoder comprises alternating layers of multi-head self-attention (MSA) (Vaswani et al. 2017) (Eq. 5) and multi-layer perceptron (MLP) (Eq. 6). Layer normalization (LN) (Ba et al. 2016) is applied before both MSA and MLP to reduce training time and stabilize the training process. Residual connections are incorporated after each layer to enhance overall performance.
Here, \(E\) refers to the operation of projecting patches, and \({E}_{{\text{pos}}}\) represents the positional embedding.
Here, \(z^{\prime }_{l}\) refers to the output of the MSA layer, applied following LN to the output of the (\(l-1\)) -th layer, denoted as \({z}_{l-1}\) (i.e.,\({z}_{0}\)), and includes a residual connection. The symbol \(L\) represents the overall number of layers in the transformer.
Here, \({z}_{l}\) refers the output of the MLP layer, applied following LN to the \(z^{\prime }_{l}\) obtained from the (\(l-1\))-th layer, and it includes a residual connection. The symbol \(L\) represents the overall number of layers in the transformer.
There are differences in image dimension processing, computational complexity, transfer learning, and performance compared to CNN architectures. By pretraining with large-scale datasets and using transfer learning, vision transformers can achieve successful results in various computer vision tasks. Additionally, it allows for the processing of large-sized images and can be more efficient in terms of computational cost due to the absence of convolution operations. The typical algorithm for ViT is presented in Algorithm 2. Furthermore, there are different architectures such as patch-based, hybrid, token-based, scale-specific, and mobile vision transformers, each offering different approaches to improve performance and efficiency.
Deep learning models
In this study, the integration of a diverse set of cutting-edge models, encompassing 14 convolutional neural network (CNN) architectures and 17 vision transformer (ViT) models, serves a strategic purpose to advance the accuracy of grape leaf classification and disease diagnosis. The selection process involved prominent CNN models such as ResNet, DenseNet, and EfficientNet, which are widely recognized for their effectiveness in image-related tasks and continue to be popular choices. Additionally, we incorporated vision transformer models such as DeiT, MaxViT, and MobileViT, which have gained popularity in recent years for their success in sequential tasks.
The rationale behind utilizing this array of models lies in their distinct architectural features and proven efficacy in various image-related tasks. CNNs, with their adeptness at capturing spatial hierarchies, are well-suited for tasks like leaf classification. On the other hand, vision transformers offer a novel perspective with their success in sequential tasks, presenting a valuable alternative for image analysis. By employing a variety of CNN and ViT models, we aim to explore and leverage the strengths of different architectures, fostering a comprehensive evaluation of their performance in the specific context of grape leaf classification and disease diagnosis.
This diverse selection is essential for understanding the nuanced capabilities of each model and collectively enhancing the robustness and generalization capacities of our approach. Through this methodology, we strive to contribute to the advancement of accuracy in grape-related tasks using deep learning techniques.
MobileVit is a version of vision transformer developed by Mehta and Rastegari (2021) called mobile vision transformer. It is designed to be run on mobile devices and enables more efficient computation. It achieves this by reducing the number of parameters and using more efficient convolution layers and other techniques.
DeiT, also known as data-efficient image transformer, is another type of vision transformer. It is specifically designed for image processing tasks and has the ability to achieve good performance with less training data compared to other vision transformer architectures. DeiT further enhances the model's performance and efficiency using techniques such as augmentation, interpolation, and distillation (Touvron et al. 2021).
Multi-axes vision transformer (MaxViT) is a type of vision transformer developed by Tu et al. (2022). It is designed to improve performance in image recognition tasks. It aims to capture multi-level representations of the image by using multiple axes. Each axis is trained to attend to different levels of image features. Additionally, it is designed to be more efficient in terms of computation compared to other vision transformer architectures.
Multiscale vision transformers (MViT) are transformer structures introduced by Fan et al. (2021) for modeling visual data such as images and videos. Compared to traditional transformers, MViT uses channel resolution scaling stages to build a feature pyramid at multiple scales. These stages reduce the spatial resolution of the image while hierarchically expanding the channel capacity. As a result, MViT can capture both low-level visual data and complex high-dimensional features using transformer principles, providing a useful feature for visual data modeling.
VGG is a CNN architecture developed by the Visual Geometry Group at the University of Oxford. This architecture has a series of convolutional and maximum pooling layers followed by three fully connected classification layers. The VGG network, designed by Simonyan and Zisserman, is developed for image detection and classification tasks. VGG's exceptional performance has been demonstrated in many image classification datasets, including ImageNet, and has been widely used as a basis for other CNN models.
ResNet is a CNN architecture that uses residual connections, known as skip connections, to overcome the vanishing gradient problem in deep networks. These connections allow the network to learn residual functions and facilitate the propagation of gradients throughout the network. ResNet has become a popular architecture in the field of computer vision, demonstrating exceptional performance in ImageNet and various other image classification tasks.
A CNN design called DenseNet was developed to overcome the vanishing gradient issue in deep neural networks. It creates close connections between layers, allowing each layer to broadcast its own feature maps to all following layers as well as receive feature maps from all preceding ones. This dense connectivity structure enables consistent feature propagation throughout the network and efficient parameter sharing. DenseNet has exhibited outstanding performance in various image classification benchmarks and has been widely used in various computer vision applications.
Xception (Chollet 2017) is a CNN design that is an extension of the Inception architecture. In Xception, instead of traditional convolutional layers, a depthwise separable convolution is used, which splits the convolution stage into a depthwise convolution and a pointwise convolution. This strategy significantly reduces the number of parameters and computations required by the network while increasing accuracy. Xception has achieved impressive results in various image classification benchmarks and has been widely used in computer vision applications.
Proposed approach
The basic components of the grape leaf disease detection and classification study are shown in Fig. 5. The first stage of this study is the Dataset unit, where two separate datasets are used. The data processing stage is concerned with resizing, splitting, and applying basic data augmentation techniques to the images in the dataset.
In the studies conducted in the literature using deep learning models, it is observed that they are trained in the train and validation steps. However, in this study, unlike the studies in the literature, the data was divided into train (15%), test (15%), and validation (70%). The train set is the dataset used for model training and optimizing its parameters. The validation set, on the other hand, is used for adjusting the hyperparameters and evaluating the overall performance of the model (Chauhan et al. 2021). However, evaluating the model based only on the train and validation sets may have limitations in predicting how the model will generalize to real-world data.
Therefore, it is important to use a test set. The test set is an independent dataset that has not been used during the training process and is used to obtain results closest to real-world data. By using a test set, it prevents the model from producing incorrect results due to its familiarity with the train set. The performance of the model is evaluated on the test set, and its success in real-world scenarios is observed. This allows for a more reliable evaluation of the overall performance and generalization ability of the model.
By using the train, test, and validation sets, the training process of the model can be optimized, overfitting can be controlled, and the real-world performance can be predicted more reliably. This differentiation is important in achieving reliable results and understanding how the model will respond to real-world data. The train, test, and validation splits of the PlantVillage dataset are presented in Table 1, and the Grapevine dataset splits are shown in Table 2.
In order to balance the class distributions, basic data augmentation techniques such as flip, rotate, copy-paste, scale and zoom are applied to the dataset during training. These data augmentation techniques are particularly effective in small-scale datasets with low diversity, while their impact is less pronounced in large-scale datasets.
In the training unit, CNN and ViT models are used for transfer learning and classification. Transfer learning refers to the process of using the weights of a model trained in one domain for a different domain. In this study, the weights of CNN and ViT models trained on the ImageNet dataset are used for disease detection in the PlantVillage dataset, and trained on the Grapevine dataset are used for class detection in the Grapevine dataset. Transfer learning is especially beneficial in small-scale datasets, and in this study, it has been observed that transfer learning leads to faster convergence and improved performance compared to training from scratch.
Following transfer learning, 14 CNN models and 17 ViT models undergo validation and testing processes for the classification task. The models are evaluated on the Test Data to derive their performance metrics. Subsequently, the model outputs and performance metrics are analyzed to execute the classification process.
Data processing
Deep learning requires data preprocessing to improve model performance by solving issues such as missing value handling and variable transformation for numerical-focused algorithms. Before feeding the data into the deep learning model, we normalized the pixel values of the images to a standardized range. Normalization ensures that the model converges more quickly during training and is less sensitive to variations in input data (Thukral et al. 2023). For consistency in input dimensions, a critical step involved resizing all images to a predefined resolution. While the majority adhered to the standard 224 × 224 resolution, it's noteworthy that specific models, such as SwinV2 (Liu et al. 2022) and Inception-based, utilized a resolution of 256 × 256 and 299 × 299, respectively. This step is crucial for ensuring that the model can handle images of uniform size, facilitating the learning process.
For reproducibility, unlike existing studies in the literature, we divided both public datasets randomly into three distinct sets: train, validation, and test (70–15-15). This unique approach ensures that each deep learning model is solely evaluated on the test data, representing unseen information, to reveal their true generalization capabilities. Tables 1 and 2 display the image counts for each class in every dataset, indicating the number of images in the train-validation-test sets.
Data augmentation
Data augmentation adds variation and variability to the training dataset, which is essential for improving the performance of machine learning models. It increases model robustness, decreases overfitting, and enhances generalization capabilities through transformations like rotation and scaling, which increases training efficiency.
Flip augmentation involves horizontally or vertically flipping the image to create new data samples. This helps the model learn how objects appear from different perspectives and aids in generalization. Rotate augmentation aims to generate various perspectives by rotating the image at a specific degree. This is particularly useful for models that need to understand the rotated appearance of objects. Copy-paste augmentation creates new examples by copying specific regions from one image and pasting them onto others. This method is valuable for understanding how objects change within different contexts. Scale augmentation generates new examples by resizing the image, either enlarging or reducing it. This is employed to learn how objects appear in different sizes and help the model generalize this diversity. Zoom augmentation creates new examples by zooming in or out of the image. This technique assists the model in understanding how objects appear up close or from a distance.
These techniques were carefully chosen based on empirical observations during the experimentation phase, aiming to strike a balance between introducing diversity and preserving the meaningful characteristics of the images (Karaman et al. 2023). By providing these specific details, we aim to offer transparency in our methodology, enabling readers to better understand and replicate the study if needed.
Results and discussions
Experimental design
This study was conducted on a Linux computer running Ubuntu 22.04, equipped with an Intel Core i5 13600 K processor, 32 GB DDR5 RAM, and an NVIDIA RTX 3090 graphics card. The experiments, performed using PyTorch with NVIDIA CUDA support, maintained a consistent environment for training and testing all architectures, each trained with the same parameters.
Performance metrics
Performance metrics play an important role in evaluating the effectiveness of deep learning models and making informed decisions. These metrics are crucial for assessing model performance, guiding the optimization process, reporting results, detecting biases and errors, making comparisons, and identifying overfitting. In this study, we specifically focused on criteria that are effective in grape disease identification and used commonly employed metrics in the literature.
Accuracy, precision, recall, and F1 score are widely used metrics in deep learning. Accuracy is defined as the ratio of correct predictions to total predictions and generally indicates the overall performance of the model. Precision measures the ratio of true positive predictions to all positive predictions and estimates the proportion of successful predictions made by the model. Recall measures the ratio of true positive predictions to all positive examples and shows how well the model identifies true positive examples. F1 score balances recall and precision and is defined as the harmonic mean of recall and precision. It is used to balance precision and recall. Each metric has a mathematical formula given by the following equations:
Training procedure
During training, deep learning models' accuracy and speed can be improved by using a variety of parameters and methodologies. The two most efficient methods are data augmentation and transfer learning. The performance of the model can also be influenced by other factors, including the input picture size, batch size, number of epochs, optimization method, learning rate, weight regularization, decay rate, and augmentation repetition. Each model was trained once with default hyperparameters, and after being saved based on the best performance on the validation set, the generalization capabilities of the models were assessed using the test data.
To assure consistency and improve performance, all models in this study included core data augmentation techniques such as scaling, smoothing, shuffling, color irregularity, and flipping. The weights from the ImageNet dataset were used to hasten convergence and boost accuracy through the use of transfer learning. Most models used training and validation data with a 224 × 224 default input resolution. All models were run using the following default parameters: lr = 0.000001, lr_base = 0.1, momentum = 0.9, optimizer = SGD, weight_decay = 2.0e-05, warmup epoch = 5, warmup lr = 1.0e-05.
Results
In this section, we present the results obtained from both CNN and vision transformer models on two distinct datasets: PlantVillage and Grapevine. These datasets encompass a wide range of grape leaf diseases and varieties, providing a comprehensive evaluation of the models' performance in different agricultural contexts.
Data processing has traditionally involved choosing between training-test, training-validation, or cross-validation methods, which may not fully demonstrate the models' performance on unseen data. In contrast to the literature, this study implemented a training-test-validation split, randomly segregating the datasets into training-validation and test sets. Consequently, each model was exclusively evaluated using the unseen test set, measuring the models' generalization capabilities. Each model underwent training once, and the best model was selected based on the validation data. Subsequently, this model was exclusively assessed on the test data, representing its performance on unseen data.
The CNN models were trained and fine-tuned on the PlantVillage dataset, which consists of various grape diseases, including Black Rot, Leaf Blight, Healthy, and Esca leaves. The evaluation of these models on the PlantVillage dataset demonstrated their ability to accurately distinguish between different grape diseases, achieving high levels of accuracy, precision, recall, and F1-score. The results showcased the effectiveness of CNN models in detecting and classifying grape leaf diseases, providing valuable insights for disease management and prevention in vineyards.
In addition to CNN models, we also evaluated vision transformer models on the Grapevine dataset. This dataset includes a diverse range of grape leaf varieties, such as Ak, Alaidris, Buzgulu, Dimnit, and Nazli. The vision transformer models, leveraging self-attention mechanisms and advanced neural network architectures, exhibited remarkable performance in accurately recognizing and classifying the grape leaf varieties. The results revealed high accuracy, precision, recall, and F1-score for the vision transformer models, indicating their potential for reliable grape leaf recognition and characterization.
Results for the PlantVillage dataset
In this section, we conducted a thorough comparison between the widely used convolutional neural network (CNN) architecture and the emerging vision transformer (ViT)-based models. The objective was to assess their performance and effectiveness in plant disease recognition and classification using the PlantVillage dataset. The comparison results are presented in Table 3 for the CNN models and Table 4 for the ViT models.
The CNN models evaluated on the PlantVillage dataset demonstrated impressive accuracy in accurately identifying and classifying grape leaf diseases. Models such as VGG-13, VGG-16, VGG-19, ResNet-18, ResNet-34, ResNet-50, Xception, Inception-v4, EfficientNetV2-S/M/L, and DenseNet models achieved high accuracy scores, with results ranging from 97 to 100%. For example, VGG-13, VGG-16, and VGG-19 achieved remarkable accuracy scores of 99.67%, 100%, and 100% respectively. Similarly, the ResNet models, including ResNet-18, ResNet-34, and ResNet-50, demonstrated high accuracy scores of 99.02%, 99.18%, and 99.51% respectively. The Xception model achieved outstanding accuracy of 99.84%, while Inception-v4 achieved an accuracy score of 99.76%.
Furthermore, the EfficientNetV2 models, ranging from EfficientNetV2-S to EfficientNetV2-L, consistently achieved high accuracy scores ranging from 99.51% to 100%. DenseNet models, including DenseNet121 and DenseNet169, also demonstrated excellent accuracy scores of 100% and 99.84% respectively. These accuracy scores emphasize the capability of the CNN models to accurately classify grape leaf diseases. The high accuracy achieved by these models indicates their potential for automated systems in grape leaf disease detection and classification, providing valuable insights for efficient disease management in agriculture.
The results for vision transformer models on the PlantVillage dataset, as shown in Table 4, reveal their impressive performance in plant disease detection and classification. Several models achieved exceptional accuracy, with some reaching up to 100%. For instance, Swinv2-base-win8, DeiT3-base, MaxViT-small, and MaxViT-large attained perfect scores across all metrics, including accuracy, precision, recall, and F1-score. These results demonstrate the robust capabilities of vision transformers in accurately identifying and classifying plant diseases. Furthermore, other vision transformer models, such as Swinv2-small-win8, Mobilevit-s, Vit-base-patch16, and Vit-large-patch16, achieved high accuracy scores ranging from 99.18 to 99.84%. These models demonstrated strong performance across all metrics, including precision, recall, and F1-score. The consistently high accuracy achieved by vision transformers highlights their effectiveness in handling complex visual patterns and capturing important dependencies within the plant images.
The comparative analysis of the vision transformer models on the PlantVillage dataset suggests that these models are highly suitable for plant disease recognition tasks. In Fig. 6, the accuracy results of the models for the PlantVillage dataset are presented in the form of a dot graph. They outperformed traditional manual approaches and achieved remarkable accuracy, surpassing the 97% threshold in most cases. The self-attention mechanisms employed by vision transformers allow them to capture important visual features, enabling accurate identification and classification of plant diseases. These findings have significant implications for the agricultural industry, as vision transformer models can aid in early disease detection, prompt intervention, and efficient disease management. By leveraging the power of deep learning and advanced image processing techniques, farmers and agricultural professionals can enhance crop productivity and minimize losses. Further research is warranted to explore the generalizability of vision transformers across different datasets and plant species, paving the way for their widespread adoption in precision agriculture and sustainable crop management practices. The confusion matrices of some models that achieved the highest accuracy and the model that achieved the lowest accuracy are provided in Fig. 7. The ones with high accuracy are indicated at the top in blue color, while the ones with low accuracy are indicated at the bottom in red color.
When we examine the VGG16 model, we can see that it achieved 177 correct classifications in the black_rot class. In the esca class, it achieved 208 correct classifications, and in the healthy class, it achieved 64 correct classifications. Additionally, it obtained 162 correct classifications in the leaf_blight class. These results demonstrate the ability of the VGG16 model to accurately identify plant diseases. Upon evaluating the ResNet18 model, we observe that it achieved 174 correct classifications in the black_rot class, 205 correct classifications in the esca class, and 64 correct classifications in the healthy class. It also obtained 162 correct classifications in the leaf_blight class. These results indicate that the ResNet18 model is capable of accurately classifying plant diseases.
In the case of the MaxViT-Small-TF-224 model, we can see that it achieved 177 correct classifications in the black_rot class, 208 correct classifications in the esca class, and 64 correct classifications in the healthy class. It also obtained 162 correct classifications in the leaf_blight class. These results demonstrate the effectiveness of the MaxViT-Small-TF-224 model in accurately classifying plant diseases. Lastly, for the ViT-Tiny-Patch16-224 model, it achieved 173 correct classifications in the black_rot class, 207 correct classifications in the esca class, and 64 correct classifications in the healthy class. It also obtained 162 correct classifications in the leaf_blight class. These results indicate that the ViT-Tiny-Patch16-224 model is also effective in accurately classifying plant diseases.
All of these models have achieved high accuracy rates on the PlantVillage dataset and have shown that they can classify plant diseases with accuracy. In conclusion, both CNN and ViT-based models demonstrate high performance on the PlantVillage dataset and prove to be effective tools in the field of deep learning.
Results for the grapevine dataset
In this section, we conducted a comprehensive comparison between the popular convolutional neural network (CNN) architecture, which is regularly used in deep learning models, and the emerging vision transformer (ViT)-based models. The objective was to evaluate their performance in the context of the Grapevine dataset, specifically focusing on the detection and classification of grape leaves. To conduct the comparison, we trained and tested a range of CNN models and ViT models, utilizing the Grapevine dataset. This dataset encompasses five classes for leaf recognition: Ak, Alaidris, Buzgulu, Dimnit, and Nazli. The models were fine-tuned and evaluated on this dataset to assess their accuracy and effectiveness in identifying and classifying the different grape leaf varieties. The detailed results of this comparison can be seen in Table 5 for the CNN models and Table 6 for the ViT models. These tables provide a comprehensive overview of the performance metrics achieved by each model, including accuracy, precision, recall, and F1 score. Additionally, they showcase the specific grape leaf varieties and their corresponding classification results for each model.
Among the models evaluated, VGG-13, VGG-16, and VGG-19 exhibited competitive performance with accuracy ranging from 96 to 97.33%. The ResNet models showed varying results, with ResNet-101 outperforming others with an accuracy of 98.67%. Xception and Inception-v4 also demonstrated outstanding performance, achieving accuracy scores of 98.67% and 100% respectively. EfficientNetV2 and DenseNet models consistently achieved high accuracy, ranging from 96.67 to 98.67%.
These findings suggest that the choice of deep learning architecture significantly impacts the performance of grape leaf recognition models. Models with complex architectures and the ability to capture intricate features, such as VGG, Xception, Inception-v4, EfficientNetV2, and DenseNet models, exhibited superior performance. However, ResNet-101 also demonstrated exceptional accuracy, showcasing the potential of deeper ResNet architectures.
Among the evaluated models, several ViT models achieved impressive results. Swinv2-tiny-win8, Swinv2-small-win8, MobileViT-xxs, MobileViT-xs, MobileViT-s, ViT-tiny-patch16, ViT-base-patch16, DeiT3-small, and DeiT3-medium achieved high accuracy scores, ranging from 96.67 to 100%. These models consistently demonstrated strong capabilities in accurately identifying and classifying grape leaf varieties.
ViT-small-patch32 and DeiT3-base models achieved slightly lower accuracy scores of 97.33%. However, they still exhibited competitive precision, recall, and F1-score metrics, highlighting their effectiveness in grape leaf recognition tasks. On the other hand, ViT-large-patch16, MaxViT-tiny, MaxViT-small, and MaxViT-base models showed relatively lower accuracy, ranging from 94.67 to 96%. These models achieved moderate precision, recall, and F1-scores, indicating their potential for grape leaf recognition but with room for improvement.
Overall, these models demonstrated high accuracies and exhibited strong classification performance on the grapevine dataset. In Fig. 8, the accuracy results of the models for the Grapevine dataset are presented in the form of a dot graph. However, it's important to consider other factors such as computational efficiency and model complexity when selecting a model for specific applications.
The confusion matrices of some models that achieved the highest accuracy and the model that achieved the lowest accuracy on Grapevine dataset are provided in Fig. 9. The ones with high accuracy are indicated at the top in blue color, while the ones with low accuracy are indicated at the bottom in red color.
Among the CNN-based models, only the Inception_v4 model achieved 15 correct classifications for the Ak, Ala_Idris, Buzgulu, Dimnit, and Nazli classes. In other words, all classes were correctly recognized. The Resnet50 model, which showed the worst performance among the CNN-based models, achieved 13 correct classifications for the Ak class, 11 for the Ala_Idris class, 13 for the Buzgulu class, 15 for the Dimnit class, and 12 for the Nazli class. Some classes also had misclassifications.
Among the ViT-based models, only the SwinV2_base_win8 model achieved 15 correct classifications for all classes. In other words, all classes were correctly recognized. The MaxViT_tiny model, which showed the worst performance among the ViT-based models, achieved 15 correct classifications for the Ak class, 14 for the Ala_Idris class, 14 for the Buzgulu class, 15 for the Dimnit class, and 13 for the Nazli class. Some classes also had misclassifications.
These results demonstrate that the ViT-based models generally performed well on the Grapevine dataset. The SwinV2_base_win8 model had the highest correct classification rates among the models we examined. It is worth noting that this model also yielded excellent results on the PlantVillage dataset. However, the other models also achieved successful results overall.
Comparison with SOTA methods
State-of-the-art (SOTA) approaches in the area were compared to the performance of the proposed deep learning-based strategy for grape leaf disease detection and classification. The results showcased the potential of our approach in achieving competitive results and advancing the current understanding of grape leaf recognition is depicted in Table 7.
Table 7 provides a comparison of the proposed approach with state-of-the-art (SOTA) methods in grape leaf recognition and classification. The comparison includes the authors, year of publication, method used, dataset employed, and the accuracy and F1-score achieved by each method.
Koklu et al. (2022) employed a combination of CNN and SVM techniques on the Grapevine dataset, achieving an accuracy and F1-score of 97.60%. This study represents one of the SOTA methods for grape leaf recognition on the Grapevine dataset. In contrast, the proposed approach in 2023 utilized a combination of CNN and ViT models on the same Grapevine dataset, achieving a perfect accuracy and F1-score of 100%. This signifies a significant advancement in grape leaf recognition performance compared to previous methods.
Regarding the PlantVillage dataset, Rao et al. (2021) utilized CNN-based models and achieved an accuracy of 99.03%. Adeel et al. (2019) employed SVM techniques and achieved an accuracy and F1-score of 97.80% and 97.62%, respectively. Yeswanth et al. (2023) and Tang et al. (2020) both utilized CNN models and achieved accuracies of 99.37% and 99.01%, respectively. In comparison, the proposed approach in 2023, using CNN models on the PlantVillage dataset, achieved a perfect accuracy and F1-score of 100%. This outperforms all the previous SOTA methods on this dataset, highlighting the superiority of the proposed approach in grape leaf recognition. Overall, the proposed approach utilizing a combination of CNN and ViT models demonstrates exceptional performance, achieving perfect accuracy and F1-score on both the Grapevine and PlantVillage datasets. This showcases the effectiveness and superiority of the proposed approach over the existing state-of-the-art methods in grape leaf recognition tasks.
Limitations and future directions
While our study has made significant strides in the application of deep learning for grape leaf classification and disease diagnosis, acknowledging certain limitations is crucial. Firstly, relying on digital images for disease detection may not fully capture the dynamic and evolving nature of plant diseases in real-world agricultural environments. Additionally, although the PlantVillage dataset is comprehensive, it may not encompass the entire spectrum of potential variations in grape diseases. Moreover, the relatively small size of the Grapevine dataset could impact the measurement of generalizability. The potential limitation of generalizing our models to different environmental conditions and grape varieties needs consideration. Furthermore, the study's focus on specific diseases and leaf types may limit its applicability to a broader range of plant-related issues.
To address these limitations and advance the field, future research should explore the integration of multimodal data, including larger datasets and additional sources such as different locations, environmental factors, and weather patterns. This holistic approach has the potential to enhance the robustness and real-world applicability of disease detection models. Collaboration with plant pathology and agronomy experts can provide valuable insights for model improvement and result validation. Future studies may broaden the scope beyond grapes by investigating the transferability of models to different crop types. The inclusion of real-time monitoring and advanced sensors could facilitate the development of more sensitive and adaptable systems for early disease detection in the field.
Conclusion
This study illuminates the transformative impact of deep learning techniques on addressing the complex challenges posed by plant diseases in agricultural contexts. Traditional approaches, rooted in expert knowledge, are often limited by inherent biases and subjectivity. Leveraging the power of computing and image processing advancements, our research delves into grape leaf classification and disease diagnosis using a combination of 14 CNN and 17 vision transformer models. The notable success of models like Swinv2-Base, achieving perfect accuracy on both the PlantVillage and Grapevine datasets, underscores the potential of deep learning in accurate disease detection and classification. This breakthrough not only marks a significant stride in bolstering crop productivity through early disease detection but also introduces a promising avenue for characterizing various grape varieties.
Looking beyond the technical achievements, the practical applications of our study resonate deeply within the agricultural sector. Our models offer a transformative potential for farmers and practitioners, facilitating informed decision-making, optimizing production efficiency, and fortifying sustainable agricultural practices. By harnessing the synergy of digital imagery and computational algorithms, our approach enables timely identification and intervention in plant diseases, mitigating production losses and fostering a more resilient and sustainable agricultural landscape. As we advance into an era where the convergence of technology and agriculture becomes increasingly vital, this research stands as a testament to the potential of deep learning to revolutionize the way we approach plant health and agricultural sustainability.
References
Abbasgholipour M, Omid M, Keyhani A, Mohtasebi SS (2011) Color image segmentation with genetic algorithm in a raisin sorting system based on machine vision in variable conditions. Expert Syst Appl 38(4):3671–3678
Adeel A, Khan MA, Sharif M, Azam F, Shah JH, Umer T, Wan S (2019) Diagnosis and recognition of grape leaf diseases: an automated system based on a novel saliency approach and canonical correlation analysis based multiple features fusion. Sustain Comput: Inf Syst 24:100349
Aggarwal AK (2020) Fusion and enhancement techniques for processing of multispectral images. Unmanned aerial vehicle: applications in agriculture and environment. Springer, Cham, pp 159–175
Aggarwal AK, Jaidka P (2022) Segmentation of crop images for crop yield prediction. Int. J. Biol. Biomed. 7:40–44
Akbarzadeh S, Paap A, Ahderom S, Apopei B, Alameh K (2018) Plant discrimination by support vector machine classifier based on spectral reflectance. Comput Electron Agric 148:250–258
Armijo G, Schlechter R, Agurto M, Muñoz D, Nuñez C, Arce-Johnson P (2016) Grapevine pathogenic microorganisms: understanding infection strategies and host response scenarios. Front Plant Sci 7:382
Ba JL, Kiros JR, Hinton G (2016) Layer normalization. arXiv preprint arXiv:1607.06450
Beikmohammadi A and Faez K (2018) Leaf classification for plant recognition with deep transfer learning. In: 2018 4th Iranian conference on signal processing and intelligent systems (ICSPIS). IEEE. pp 21–26.
Boissard P, Martin V, Moisan S (2008) A cognitive vision approach to early pest detection in greenhouse crops. Comput Electron Agric 62(2):81–93
Chauhan S, Singh M, Aggarwal AK (2021) Experimental analysis of effect of tuning parameters on the performance of diversity-driven multi-parent evolutionary algorithm. In: 2021 IEEE 2nd international conference on electrical power and energy systems (ICEPES). IEEE. pp 1–6
Chollet, F (2017) Xception: deep learning with depthwise separable convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp 1251–1258
Coşkun D, Karaboğa D, Baştürk A, Akay B, Nalbantoğlu ÖU, Doğan S, Karagöz MA (2023) A comparative study of YOLO models and a transformer-based YOLOv5 model for mass detection in mammograms. Turk J Electr Eng Comput Sci 31(7):1294–1313
Eli-Chukwu NC (2019) Applications of artificial intelligence in agriculture: a review. Eng, Technol Appl Sci Res 9(4):4377–4383
Fan H, Xiong B, Mangalam K, Li Y, Yan Z, Malik J, and Feichtenhofer C (2021) Multiscale vision transformers. In Proceedings of the IEEE/CVF International Conference on Computer Vision. pp 6824–6835
Ferentinos KP (2018) Deep learning models for plant disease detection and diagnosis. Comput Electron Agric 145:311–318
Fuentes A, Yoon S, Kim SC, Park DS (2017) A robust deep-learning-based detector for real-time tomato plant diseases and pests recognition. Sensors 17(9):2022
Ghoury S, Sungur C, and Durdu A (2019) Real-time diseases detection of grape and grape leaves using faster r-cnn and ssd mobilenet architectures. In International conference on advanced technologies, computer engineering and science (ICATCES 2019). pp. 39–44
Hamuda E, Mc Ginley B, Glavin M, Jones E (2017) Automatic crop detection under field conditions using the HSV colour space and morphological operations. Comput Electron Agric 133:97–107
Hekimoğlu B, and Altındeğer M (2006) Organik Tarım ve Bitki Koruma Açısından Organik Tarımda Kullanılacak Yöntemler. Samsun Valiliği Gıda Tarım Ve Hayvancılık İl Müdürlüğü.
Hinton GE, Osindero S, Teh YW (2006) A fast learning algorithm for deep belief nets. Neural Comput 18(7):1527–1554
Hughes D, and Salathé M (2015) An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv preprint arXiv:1511.08060
Ji M, Zhang L, Wu Q (2020) Automatic grape leaf diseases identification via united model based on multiple convolutional neural networks. Inf Process Agric 7(3):418–426
Jin H, Li Y, Qi J, Feng J, Tian D, Mu W (2022) GrapeGAN: Unsupervised image enhancement for improved grape leaf disease recognition. Comput Electron Agric 198:107055
Karaman A, Pacal I, Basturk A, Akay B, Nalbantoglu U, Coskun S, Karaboga D (2023) Robust real-time polyp detection system design based on YOLO algorithms by optimizing activation functions and hyper-parameters with artificial bee colony (ABC). Expert Syst Appl 221:119741
Kaya E, Saritas İ (2019) Towards a real-time sorting system: identification of vitreous durum wheat kernels using ANN based on their morphological, colour, wavelet and gaborlet features. Comput Electron Agric 166:105016
Kiliçarslan S, Pacal I (2023) Domates Yapraklarında Hastalık Tespiti İçin Transfer Öğrenme Metotlarının Kullanılması. Mühendislik Bilimleri Ve Araştırmaları Dergisi 5(2):215–222
Koklu M, Ozkan IA (2020) Multiclass classification of dry beans using computer vision and machine learning techniques. Comput Electron Agric 174:105507
Koklu M, Unlersen MF, Ozkan IA, Aslan MF, Sabanci K (2022) A CNN-SVM study based on selected deep features for grapevine leaves classification. Measurement 188:110425
Kulin M, Kazaz T, Moerman I, De Poorter E (2018) End-to-end learning from spectrum data: A deep learning approach for wireless signal identification in spectrum monitoring applications. IEEE Access 6:18484–18501
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444. https://doi.org/10.1038/nature14539
Li G, Ma Z, and Wang H (2012) Image recognition of grape downy mildew and grape powdery mildew based on support vector machine. In: Computer and Computing Technologies in Agriculture V: 5th IFIP TC 5/SIG 5.1 Conference, CCTA 2011, Beijing, China, October 29-31, 2011, Proceedings, Part III. Springer Berlin Heidelberg Vol 5, pp. 151–162
Liu B, Tan C, Li S, He J, Wang H (2020) A data augmentation method based on generative adversarial networks for grape leaf disease identification. IEEE Access 8:102188–102198
Liu Z, Hu H, Lin Y, Yao Z, Xie Z, Wei Y, Ning J, Cao Y, Zhang Z, Dong L, Wei F, Guo B (2022) Swin transformer v2: Scaling up capacity and resolution. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, New Orleans, LA, USA. pp 12009–12019
Manavalan R (2020) Automatic identification of diseases in grains crops through computational approaches: a review. Comput Electron Agric 178:105802
Martinelli F, Scalenghe R, Davino S, Panno S, Scuderi G, Ruisi P, Dandekar AM (2015) Advanced methods of plant disease detection. A review. Agron Sustain Dev 35:1–25
Mehta S and Rastegari M (2021) Mobilevit: light-weight, general-purpose, and mobile-friendly vision transformer. arXiv preprint arXiv:2110.02178
Mishra PK, Maurya SK, Singh RK, Misra AK (2012) A semi automatic plant identification based on digital leaf and flower images. In: IEEE-international conference on advances in engineering, science and management (ICAESM-2012). IEEE. pp 68–73
Mohammadpoor M, Nooghabi MG, and Ahmedi Z (2020) An intelligent technique for grape fanleaf virus detection
Murakami S, Homma K, and Koike T (2005) Detection of small pests on vegetable leaves using GLCM. In: 2005 ASAE Annual Meeting. American Society of Agricultural and Biological Engineers. p 1
Muthukannan K, Latha P (2018) A GA_FFNN algorithm applied for classification in diseased plant leaf system. Multimed Tools Appl 77(18):24387–24403
Pacal I (2024a) MaxCerVixT: a novel lightweight vision transformer-based approach for precise cervical cancer detection. Knowledge-Based Systems 289:111482
Pacal I (2024b) Enhancing crop productivity and sustainability through disease identification in maize leaves: exploiting a large dataset with an advanced vision transformer model. Expert Syst Appl 238:122099
Pacal I, Kılıcarslan S (2023) Deep learning-based approaches for robust classification of cervical cancer. Neural Comput Appl 35(25):18813–18828
Patrício DI, Rieder R (2018) Computer vision and artificial intelligence in precision agriculture for grain crops: a systematic review. Comput Electron Agric 153:69–81
Paulus I, De Busscher R, Schrevens E (1997) Use of image analysis to investigate human quality classification of apples. J Agric Eng Res 68(4):341–353
Peng Y, Zhao S, Liu J (2021) Fused deep features-based grape varieties identification using support vector machine. Agriculture 11(9):869
Pydipati R, Burks TF, Lee WS (2006) Identification of citrus disease using color texture features and discriminant analysis. Comput Electron Agric 52(1–2):49–59
Rao US, Swathi R, Sanjana V, Arpitha L, Chandrasekhar K, Naik PK (2021) Deep learning precision farming: grapes and mango leaf disease detection by transfer learning. GlobTrans Proc 2(2):535–544
Sandhu H (2021) Bottom-up transformation of agriculture and food systems. Sustainability 13(4):2171
Shariff AR, Aik YY, Hong WT, Mansor S, Mispan R (2006) Automated identification and counting of pests in the paddy fields using image analysis. In: Computers in agriculture and natural resources, 23–25 July 2006, Orlando Florida. American Society of Agricultural and Biological Engineers. p 759
Singh V, Misra AK (2017) Detection of plant leaf diseases using image segmentation and soft computing techniques. Inf Process Agric 4(1):41–49
Tang Z, Yang J, Li Z, Qi F (2020) Grape disease image classification based on lightweight convolution neural networks and channelwise attention. Comput Electron Agric 178:105735
Tavakoli H, Alirezazadeh P, Hedayatipour A, Nasib AB, Landwehr N (2021) Leaf image-based classification of some common bean cultivars using discriminative convolutional neural networks. Comput Electron Agric 181:105935
Thukral R, Aggarwal AK, Arora AS, Dora T, Sancheti S (2023) Artificial intelligence-based prediction of oral mucositis in patients with head-and-neck cancer: a prospective observational study utilizing a thermographic approach. Cancer Res, Stat Treat 6(2):181–190
Touvron H, Cord M, Douze M, Massa F, Sablayrolles A, Jégou H (2021) Training data-efficient image transformers & distillation through attention. In International conference on machine learning. PMLR. pp 10347–10357
Tu Z, Talebi H, Zhang H, Yang F, Milanfar P, Bovik A, Li Y (2022) Maxvit: multi-axis vision transformer. In: Computer Vision–ECCV 2022: 17th European Conference, Tel Aviv, Israel, October 23–27, 2022, Proceedings, Part XXIV. Cham: Springer Nature Switzerland. pp 459–479
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I (2017). Attention is all you need. Advances in neural information processing systems. Vol 30
Veziroglu E, Pacal I, Coşkunçay A (2023) Derin Evrişimli Sinir Ağları Kullanılarak Pirinç Hastalıklarının Sınıflandırılması. J Inst Sci Technol 13(2):792–814
Waghmare H, Kokare R, and Dandawate, Y (2016) Detection and classification of diseases of grape plant using opposite colour local binary pattern feature and machine learning for automated decision support system. In: 2016 3rd international conference on signal processing and integrated networks (SPIN). IEEE. pp. 513–518
Xiao J, Aggarwal AK, Rage UK, Katiyar V, Avtar R (2023) Deep learning-based spatiotemporal fusion of unmanned aerial vehicle and satellite reflectance images for crop monitoring. IEEE Access. https://doi.org/10.1109/ACCESS.2023.3297513
Yeswanth PV, Deivalakshmi S, George S, Ko SB (2023) Residual skip network-based super-resolution for leaf disease detection of grape plant. Circuits, Syst Signal Process 42(11):1–29
Zhang S, Wang Z (2016) Cucumber disease recognition based on Global-Local Singular value decomposition. Neurocomputing 205:341–348
Zhang Y, Gravina R, Lu H, Villari M, Fortino G (2018) PEA: Parallel electrocardiogram-based authentication for smart healthcare systems. J Netw Comput Appl 117:10–16
Zhu J, Wu A, Wang X, Zhang H (2020) Identification of grape diseases using image analysis and BP neural networks. Multimed Tools Appl 79:14539–14551
Funding
Open access funding provided by the Scientific and Technological Research Council of Türkiye (TÜBİTAK).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors affirm that they do not have any Conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kunduracioglu, I., Pacal, I. Advancements in deep learning for accurate classification of grape leaves and diagnosis of grape diseases. J Plant Dis Prot 131, 1061–1080 (2024). https://doi.org/10.1007/s41348-024-00896-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41348-024-00896-z