Abstract
The fall armyworm (FAW), an invasive pest of maize, is an emerging threat in Southern Asia after America and Europe. Recently, this notorious pest has also been found in different areas of Pakistan. To assess its presence in Pakistan, a survey was carried out in the provinces of Punjab, Sindh, and Khyber Pakhtunkhwa during May–October 2019. We observed the highest incidence of FAW in Sindh with maximum impact in districts Tando-Allahyar and Hyderabad. These samples were identified as Spodoptera frugiperda on the morphological and taxonomical bases. However, morphological identification of this pest is very difficult at early larval instars. Here, we use the mitochondrial cytochrome c oxidase I (COI) gene region for the precise identification of larva of this invasive pest at species level. Two different regions of COI gene (COI-5′ and COI-3′) were used as molecular markers for the identification of this species. DNA sequence similarity searches of the obtained COI gene sequences (NCBI GenBank Accession Nos. MW241537, MW241538, MW349515, MW349516, MW349493 and MW349494) revealed that genetically it is more than 99% identical to S. frugiperda. The phylogenetic analysis indicated it as the rice-strain (R-strain). Both 3′- and 5′-fragment tree topologies showed that the collected samples of the FAW species belong to the R-strain. To the best of our knowledge, this is the first report providing molecular evidence for the existence of R-strain of S. frugiperda that was found feeding on maize crop in Sindh, Pakistan, using COI gene sequences as a marker.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The fall armyworm (FAW) Spodoptera frugiperda J.E. Smith (Lepidoptera; Noctuidae) is a pest of many crops. Like all other noctuids, it is a polyphagous pest but rapacious feeder of cultivated grasses, especially maize. Various level of damage has been reported on different crops depending upon the phonological stage of crop (Goergen et al. 2016; FAO and CABI 2019). However, in maize that is the most preferred host of FAW, the adult larvae developed into cutworms owing to their much stronger mandibles having serrated cutting edges that can feed much easily on leaves (Goergen et al. 2016). Moreover, definitely this pest may have varietal preferences; however, the varied level of infestation may be due to climate changes of different geographical conditions. Recently, it has been spread dramatically from tropical and subtropical regions of the America to other continents representing it as the emerging threat of invasive pest species (Richardson et al. 2020). This pest has frequently been intercepted at quarantine in Europe and Africa (Goergen et al. 2016; Day et al. 2017; Rwomushana et al. 2018). Its rapid spread is causing severe damage to maize since 2016 in about 44 African countries with collective yield losses of about USD 13.38 billion (Goergen et al. 2016; Prasanna et al. 2018; IITA 2016; Rwomushana et al. 2018). FAW has more than 350 host plants, including rice, sorghum, sugarcane, and some vegetables but its preferred host is maize and around 98% farms have been reported severely damaged by FAW inflicting about 40–70% yield losses (Wyckhuys and O’Neil 2006; Day 2017). However, in the Asian subcontinent, it was first reported in 2018 from India (Shylesha et al. 2018; Ganiger et al. 2018; Sharanabasappa et al. 2018) with later incidences in Sri Lanka, Nepal, Bangladesh, China, and Pakistan (Naeem-Ullah et al. 2019; FAO, 2019a. Furthermore, looking at the wide range of hosts and owing to fast dispersing abilities with a flight range of about 100 km/night from the place of incidence (FAO 2019b, the bordering countries are likely to be attacked with increasing infestation by FAW (Naeem-Ullah et al. 2019). The agroclimate of Pakistan is very similar to India and being the neighboring country, its presence has already been reported in the maize growing districts of Upper Sindh (Naeem-Ullah et al. 2019; Gilal et al. 2020). Recently, a report confirming the presence of S. frugiperda on maize from Faisalabad has also been published (Khan et al. 2020), and the captured samples from these areas were identified on morphological characters alone. However, the confirmation of species on molecular level was required to ensure its proper identification and possible routes of entry to the maize growing areas of Pakistan.
In the present scenario of climate change and huge habitat destruction, the reliable identification of species is a pivotal component for biodiversity studies but given the magnitude of arthropod diverseness, conventional and morphological identification of larval stages associated with adult morphology and reproductive structures is a difficult task for many taxa (Phillips et al. 2019; Watts et al. 2019; Tang et al. 2010). Owing to the increased globalization, identification of invasive species through DNA barcoding is recommended using clear annotations of database records for suspected specimen origin versus interception location (Madden et al. 2019). Though, routine identification of many species at the molecular level could be difficult and time-consuming, representing a limiting factor for biodiversity assessments; however, its use as a complementary identification technique is recommended that can provide secondary evidence to support morphological identifications (Hajibabaei et al. 2007). Nevertheless, DNA barcoding is a promising and effective tool for fast and accurate species identification using precise genetic markers (Kress and Erickson 2008; Pereira et al. 2008).
The interception of any invasive pest at ports-of-entry is a crucial component of biosecurity and biosurveillance programs that requires identification of the pest species for quarantine decisions. However, the morphological identification of the Noctuid pest species of FAW is very difficult at the 1st, 2nd, and 3rd instar stages. Although at the 4th and 5th developmental instars, it can be identified on conspicuous morphological characters but by that time the economic damage reaches beyond the control for target crop. Hence, proper identification of this pest at early stage is required. The S. frugiperda consists of two strains adapted to different host-plants; the corn strain (C-strain) preferentially feeds on maize, cotton, and sorghum, whereas the rice strain (R-strain) is more associated with rice and several pasture grasses. These strains are morphologically indistinguishable; however, differences in mating behavior, pheromone compositions, host-plant adaptation, and genetic background are existing (Dumas et al. 2015). Regarding molecular characterization of FAW from Asia, only two reports from India confirmed the presence of R-strain on rice (Kalleshwaraswamy et al. 2019; Maruthadurai et al. 2020). However, some unpublished results from Pakistan exist in the GenBank database of NCBI for Accession numbers MT180095.1 to MT180098.1, showing the presence of R-strain of S. frugiperda collected from maize situated in Punjab. But this is very little information so there is a great need for more data regarding the identification of S. frugiperda at molecular level. Here, we report the first molecular characterization of FAW collected from maize crop area situated in Sindh, Pakistan. This study was carried out to survey the different areas of Pakistan (mostly likely bordering areas) for the occurrence of FAW and upgrade knowledge about the spatial distribution of FAW in Pakistan (if any). The presence of S. frugiperda was confirmed by the morphological as well as molecular analyses. Consequently, the actions like surveillance, awareness programs and capacity building measures are very crucial at this early stage of invasion.
Materials and methods
Occurrence and morphological identification
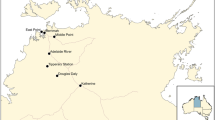
A survey for the detection and occurrence of FAW was conducted in three provinces of Pakistan, i.e., Punjab (31.1704° N, 72.7097° E), Sindh (25.8943° N, 68.5247° E) and Khyber Pakhtunkhwa (34.9526° N, 72.3311° E) where maize is grown as a major crop. A total of 32 districts were sampled for FAW occurrence (Fig. 1). The FAW presence was determined by using various indicators, i.e., (a) presence of fresh frass in the leaf funnel; (b) irregular damage on leaves (c) presence of larvae on leaves or in the leaf funnel with the inverted Y-Shape in the head and the set of four dot forming a square on the upper surface of the last segment of its body as per Prasanna et al. 2018 and (d) presence of eggs masses. The samples (larvae and adult) were identified morphologically consulting dichotomous key (Passoa 1991) and preserved in 80% ethanol for further processing of molecular identification.
FAW surveillance in three provinces of Pakistan. Where the highest incidence of FAW was observed in many districts of Sindh. In Punjab low existence of FAW was witnessed, especially in the areas that shares borders with India; however, no incidence of FAW was observed in Khyber Pakhtunkhwa (figure is
Molecular identification
DNA extraction and PCR amplification
Total genomic DNA was extracted from the collected specimen using CTAB extraction buffer (2 g of CTAB hexadecyl-trimethylammonium bromide, 10 mM Tris, 50 mM EDTA [Ethylene diaminetetraacetic acid], 0.7 M NaCl). The homogenate was incubated at 65ºC for 30 min, purified by adding an equal volume of chloroform/isoamyl alcohol mixture (24:1) and centrifugation. The upper aqueous phase (that contained insect DNA) was separated and DNA was precipitated by adding 500 μl of cold isopropanol and 10 μl of 3 M sodium acetate. The DNA pellet was collected by centrifuging the samples at 10,000 rpm for 5 min at 4 °C and washed with 70% ethanol, air-dried at room temperature and resuspended in 50 μL of TE buffer (10 mM Tris, 1 mM EDTA, pH 8). The DNA was kept at − 20ºC for long-term storage.
PCR amplification of the 3'-fragment (814 bp) of the mitochondrial gene was carried out using the primer pair C1J2195/TL2N3014 (TTGATTTTTTGGTCATCCAGAAGT/TCCAATGCACTAATCTGCCATATTA; Frohlich et al. 1999) and DreamTaq Green PCR Master Mix (Thermo Fisher Scientific). The PCR cycling parameters were one denaturation cycle of 94 °C for 5 min, followed by 35 cycles of 94 °C for 1 min, 48 °C for 1 min, and 72 °C for 1 min, followed by a final extension at 72 °C for 7 min. Similar parameters were used for the amplification of 5'-fragment (658 bp) of mitochondrial gene by LCO/HCO primers pair (GGTCAACAAATCATAAAGATATTGG/TAAAGTTCAGGGTGACCAAAAAATCA; Folmer et al. 1994).
Cloning, sequencing and sequence analysis
The PCR products were ligated in the pTZ57R/T plasmid vector (Thermo Fisher Scientific) and transformed into E.Coli Top10. The clones were confirmed by restriction analysis using EcoRI/HindIII and purified using a plasmid isolation kit (Thermo Fisher Scientific). The sequence of each cloned insert was determined by bi-directional, automated Sanger dideoxy chain-termination sequencing by Eurofins USA through Molecular Biology Products. The DNA sequences were assembled, aligned and edited using Lasergene software (DNASTAR, Madison, WI, USA). The sequences were submitted in NCBI GenBank. Sequence alignment, pairwise distance analyses and phylogenetic analyses were performed using MEGAX by using neighbor-joining and the algorithm of Maximum composite likelihood with 1000 bootstrap re-samplings (Tamura et al. 2011; Maruthadurai et al. 2020). The phylogenetic tree was generated using COI sequences from the NCBI Genbank database representative of both R and C strains of FAW.
Results
Occurrence and morphological identification
A total of 536 fields were sampled in Punjab, but only 12 fields were positive for the occurrence of FAW based on the morphological identification. The average incidence of FAW in Punjab was only 2.10%. The positive sites were located in Faisalabad and Lahore only. Regarding the occurrence of FAW in Khyber Pakhtunkhwa, a total of 55 fields were sampled; however, no field sample was found positive for the occurrence of FAW. Regarding the occurrence of FAW in Sindh (25.8943° N, 68.5247° E), a total of 338 fields were surveyed. Out of 338, 198 fields were found positive for the occurrence of FAW based on the morphological identification (Fig. 1). The average incidence of FAW in Sindh was 56.12%. The positive sites were located in the regions of TandoAllahyar (94.11%), Hyderabad (93%), Thatta (80%), Tandoadam (70%), Matiari (66.18%), Tando Muhammad Khan (58.82), Mirpurkhas (53.85%) and Umerkot (47.37%) arranged in descending order of percent occurrence in sample fields.
The key feature in full grown larva was distinct whitish to yellow colored inverted ‘Y’-shaped epicranial sutures in front of head and the larval body was covered with the elevated conspicuous spots (pinacula) along with spines with clear trapezoidal design of four pinacula on the last (9th) abdominal segment. All of the adults were copper colored with grayish-brown forewings having sidelong v-shaped marking in males, whereas forewings of females were uniformly grayish brown without any distinct markings. The morphological characters of larvae and adults were similar as reported by Maruthadurai and Ramesh (2020). Considering higher incidence of FAW in Sindh region (25.8943° N, 68.5247° E), further study is related to the larval sample collected from this region only.
Phylogenetic analysis of COI-3' and COI-5' sequences
The mtCOI-3′ sequence (814 bp) and mtCOI-5′ sequence (658 bp) are determined bi-directionally for cloned PCR amplicons. The manually edited final sequences were obtained and are available in the nucleotide sequence database under the accession numbers MW241537, MW349493 and MW349494 for mtCOI-3′. Regarding mtCOI-5′ sequences, the accession numbers are MW241538, MW349515 and MW349515.
The sequences were trimmed to remove primer sequences and BLAST aligned with mtCOI-3′ and mtCOI-5′ sequences already available in database. Using this approach, samples were identified as R-strain of fall armyworm. The identified samples showed 99% similarity with the fall armyworm samples of India. Phylogenetic tree based upon mtCOI-5′ showed that fall armyworm samples belong to the R-strain (Fig. 2). Similarly using mtCOI-3′ sequences a tree was also generated (Fig. 3) and it supported the mtCOI-5ʹ results which confirm their identity as S. frugiperda R-strain.
Pairwise distance among our samples ranges from 0.0 to 0.2%, while the pairwise distance calculated among our sequences and the sequences reported from India (MH779587, LC582517, MH779590) range from 0.0 to 0.2%.
Discussion
Previously, limited studies have been conducted on the presence of FAW in Pakistan. Most of the studies revealed the presence of this pest in different localities of Sindh province based on its morphological characteristics (Naeem-Ullah et al. 2019; Gilal et al 2020). Recently, one report also confirmed the presence of FAW in Punjab region (Khan et al. 2020) based on morphological characteristics and we still lack any report based upon molecular marker/s that confirm the presence of FAW from Pakistan.
The present study is therefore the first record of the presence of R strain of FAW in Sindh, Pakistan, using both the morphological characteristics and the advanced molecular techniques. The key morphological features of larvae and adults endorsed the existence of FAW (Todd and Poole 1980; Passoa 1991; Pogue 2002; Gilligan and Passoa 2014; Ganiger et al. 2018; Sisodiya et al. 2018; Shylesha et al. 2018; Naeem-Ullah et al. 2019; Gilal et al. 2020). The phylogenetic analysis showed that the FAW strain collected from maize fields is the R-strain of S. frugiperda. However, it has been reported that R-strain preferentially feeds on rice and various pasture grasses and the C-strain feeds on maize, cotton and sorghum (Juárez et al. 2014). Although some host-associated differentiation is present in S. frugiperda, some other factors must be maintaining genetic differentiation between the two strains (Pashley 1986). Nevertheless, we report here that the R-strain has been found feeding on the maize crop in Pakistan. Earlier, Sharanabasappa et al. (2018) have shown its presence on maize and later Kalleshwaraswamy et al. (2019) and on rice and now Maruthadurai et al. (2020) on maize in India. It seems that the R-strain is prevailing in the sub-continent as well as changing and/or expanding its host range. Now, its presence on maize in Pakistan is alarming, and its destructive nature may also damage the rice crop in the coming season, causing huge losses to crop protection in Pakistan.
The molecular identification of S. frugiperda has been reported using PCR–RFLP (Cano-Calle et al., 2015; Rosas et al. 2016), sequencing of mitochondrial gene and phylogenetic analysis (Jacobs et al., 2018; Otim et al., 2018; Maruthadurai et al. 2020). Similarly in the present study, we obtained sequences of two different regions of the COI gene (COI-5′ and COI-3′) and two phylogenetic trees were produced using sequences amplified by HCO/LCO and CO1/CO2 primers sets. Regarding the molecular identification of S. frugiperda, our results are greatly reliable and novel in respect of providing the sequence data of both N and C-terminus of COI gene. In our study, there was a wide variation in incidence/occurrence among the regions but not among the samples. Here, we showed the occurrence and molecular identification based on the COI gene; however, further studies based on PCR–RFLP may check the ratio of R- and C-strains of FAW in Pakistan if required, while the pairwise distance calculated among our sequences and other sequences ranges from 0.0 to 0.2% which showed the highest sequence similarity among samples collected from different countries. The geographical variations and the differences in the climatic conditions of sampling regions during the collection period (June–August) might be a good explanation of this variation and the differences observed. A closer depiction can be obtained on the basis of yearly data if and when available. As already reported due to the migratory nature of FAW, its presence on other host plants within similar ecological conditions may exist in the neighboring country (Kalleshwaraswamy et al. 2019), and it is a possible threat to maize crop in Pakistan. Our results are in line showing the Sindh areas sharing borders with India had a high incidence of FAW (Navik et al. 2021). Climate change can further trigger FAW outbreaks (Díaz-Álvarez et al. 2021).
Successful pest management programs rely on prior knowledge of the correct identification of pest and host preferences. Such a result regarding correct identification of existing strain is of crucial value and may be used to develop synthetic pesticides and other strategies. As the present study confirms the presence of S. frugiperda in maize growing areas of Sindh, it is suggested that the government and other key stakeholders should consider this upcoming threat to maize crop and allied industries. The pest attack by S. frugiperda could result in substantial yield losses and considerably higher costs of production attributable to reliance on chemical control. This demands careful future planning by the Government and time bound research and development activities to develop suitable IPM strategies for its control. Henceforth, some more surveys and the molecular identification of the existing species for biodiversity analysis from distant areas are also recommended.
Conclusion
R-strain of fall armyworm (FAW) has been found feeding on maize crop in Sindh, Pakistan. This is the first report providing molecular-based evidences regarding occurrence of this invasive pest on maize in Sindh, Pakistan, that has expanded its host. It may also damage to the rice and other crops in the coming season. Considering the importance of maize and rice crops in Pakistan, this study may serve as a basis for further surveys of FAW, distribution, molecular identification and implementation of IPM strategies. This identification may also be used to develop synthetic pesticides and design some genetically engineered strategies.
References
Cano-Calle D, Arango-Isaza RE, Saldamando-Benjumea CI (2015) Molecular identification of spodoptera frugiperda (Lepidoptera: Noctuidae) corn and rice strains in colombia by using a PCR-RFLP of the mitochondrial gene cytochrome oxydase I (COI) and a PCR of the Gene FR (For Rice). Ann Entomol Soc Am 108:172–180. https://doi.org/10.1093/aesa/sav001
Day R, Abrahams P, Bateman M, Beale T, Clottey V, Cock MJ, Gomez J (2017) Fall armyworm: impacts and implications for Africa. Outlooks Pest Manag 28:196–201
Díaz-Álvarez EA, Martínez-Zavaleta JP, López-Santiz EE, la Barrera E, Larsen J, del-Val E (2021) Climate change can trigger fall armyworm outbreaks: a developmental response experiment with two Mexican maize landraces. Int J Pest Manag. DOI: https://doi.org/10.1080/09670874.2020.1869347
Dumas P, Fabrice L, Claire L, Erwan S, Marion O, Le K, Sylvie G (2015) Spodoptera frugiperda (Lepidoptera: Noctuidae) host-plant variants: two host strains or two distinct species? Genetica 143:305–316. https://doi.org/10.1007/s10709-015-9829-2
FAO (2019 a) Briefing note on FAO actions on fall armyworm
FAO (2019 b) Fall armyworm (FAW); Q & A. http://www.fao.org/3/a-i7471e.pdf
FAO-CABI (2019) Community-based fall armyworm (Spodoptera frugiperda ) monitoring, early warning and management. training of trainers manual (First Edition). Food Agric. Organ. U. N. CAB Int
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299 (PMID: 7881515)
Frohlich DR, Torres-Jerez I, Bedford ID, Markham PG, Brown JK (1999) A phylogeographical analysis of Bemisia tabaci species complex based on mitochondrial DNA markers. Mol Ecol 8:1683–1691
Ganiger PC, Yeshwanth HM, Muralimohan K, Vinay N, Kumar ARV, Chandrashekara K (2018) Occurrence of the new invasive pest, fall armyworm, Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae), in the maize fields of Karnataka. India Curr Sci 115:621–623
Gilal AA, Bashir L, Faheem M, Rajput A, Soomro JA, Kunbhar S, Mirwani AS, MastoiSahito GSJGM (2020) First record of invasive fall armyworm Spodoptera frugiperda Smith (Lepidoptera: Noctuidae) in corn fields of Sindh, Pakistan. Pakistan J Agri Res 33:247–252
Gilligan TM, Passoa SC (2014) LepIntercept, an identification resource for intercepted Lepidoptera Larvae. Identification technology program (ITP), USDA-APHIS-PPQ-S&T, Fort Collins (US). www.lepintercept.org
Goergen G, Kumar PL, Sankung SB, Togola A, Tamo M (2016) First report of outbreaks of the fall armyworm Spodoptera frugiperda (JE Smith) (Lepidoptera, Noctuidae), a new alien invasive pest in west and central Africa. PLoS ONE 11(10):e0165632. https://doi.org/10.1371/journal.pone.0165632
Hajibabaei M, Singer GAC, Clare EL, Hebert PDN (2007) Design and applicability of DNA arrays and DNA barcodes in biodiversity monitoring. BMC Biol 5:24
Jacobs A, van Vuuren A, Rong IH (2018) Characterisation of the fall armyworm (Spodoptera frugiperda JE Smith) (Lepidoptera: Noctuidae) from South Africa. Afr Entomol 26:45–49. https://doi.org/10.4001/003.026.0045
Juárez ML, Schöfl G, Vera MT et al (2014) Population structure of Spodoptera frugiperda maize and rice host forms in South America: are they host strains? Entomol Exp Appl. https://doi.org/10.1111/eea.12215
Kalleshwaraswamy CM, Asokan R, Mahadevaswamy HMM (2019) First record of invasive fall armyworm, Spodoptera frugiperda (JE Smith)(Lepidoptera: Noctuidae) on rice (Oryza sativa) from India. J Entomol Zool Stud 7:332–337
Khan HA, Ali N, Farooq MU, Asif N, Gill TA, Khalique U (2020) First authentic report of fall armyworm presence in Faisalabad Pakistan. J Entomol Zool Stud 8:1512–1514
Kress WJ, Erickson DL (2008) DNA barcodes: genes, genomics, and bioinformatics. PNAS 105:2761–2762
Madden MJL, Young RG, Brown JW, Miller SE, Frewin AJ, Hanner RH (2019) Using DNA barcoding to improve invasive pest identification at US ports–of–entry. PLoS ONE 14(9):e0222291. https://doi.org/10.1371/journal.pone.0222291
Maruthadurai R, Ramesh R (2020) Occurrence, damage pattern and biology of fall armyworm, Spodoptera frugiperda (JE smith) (Lepidoptera: Noctuidae) on fodder crops and green amaranth in Goa, India. Phytoparasitica 48:15–23. https://doi.org/10.1007/s12600-019-00771-w
Naeem-Ullah U, Ansari MA, Iqbal N, Saeed S (2019) First authentic report of Spodoptera frugiperda (JE Smith) (Noctuidae: Lepidoptera) an alien invasive species from Pakistan. ASBE 6:1–3
Navik O, Shylesha AN, Patil J, Venkatesan T, Lalitha Y, Ashika TR (2021) Damage, distribution and natural enemies of invasive fall armyworm Spodoptera frugiperda (JE Smith) under rainfed maize in Karnataka India. Crop Prot 143:105536
Otim MH, Tay WT, Walsh TK, Kanyesigye D, Adumo S, Abongosi J (2018) Detection of sister-species in invasive populations of the fall armyworm Spodoptera frugiperda (Lepidoptera: Noctuidae) from Uganda. Plos One. https://doi.org/10.1371/journal.pone.0194571
Pashley DP (1986) Host associated genetic differentiation in fall armyworm (Lepidoptera: Noctuidae): a sibling species complex? Ann Entomol Soc Am 79:898–904
Passoa S (1991) Color identification of economically important Spodoptera larvae in Honduras (Lepidoptera: Noctuidae). Insecta Mundi 5:185–196
Pereira F, Carneiro J, Amorim A (2008) Identification of species with DNA-based technology: current progress and challenges. Recent Pat DNA Gene Seq 2:187–200
Phillips JD, Gillis DJ, Hanner RH (2019) Incomplete estimates of genetic diversity within species: implications for DNA barcoding. Ecol Evol 9:2996–3010. https://doi.org/10.1002/ece3.4757 (PMID: 30891232)
Pogue MG (2002) A world revision of the genus Spodoptera Guenée (Lepidoptera: Noctuidae). Mem Am Entomol Soc 43:1–202
Prasanna BM, Huesing JE, Eddy R, Peschke VM (2018) Fall armyworm in Africa: a guide for integrated pest management. 1st ed. CIMMYT; Edo Mex, Mexico. https://reliefweb.int/sites/reliefweb.int/files/resources/FallArmyworm_IPM_Guide_forAfrica.pdf.
Richardson EB, Troczka BJ, Gutbrod O (2020) Diamide resistance: 10 years of lessons from lepidopteran pests. J Pest Sci 93:911–928. https://doi.org/10.1007/s10340-020-01220-y
Rosas-Garcia NM, Herrera-Mayorga V, Rivera G, Mireles-Martinez M, Sanchez FAP, Villegas-Mendoza JM (2016) Spodoptera frugiperda biotype identification from maize host plants in different regions of Mexico. Southwest Entomol 41:761–770. https://doi.org/10.3958/059.041.0318
Rwomushana I, Bateman M, Beale T, Beseh P, Cameron K, Chiluba M and Clottey V (2018) Fall armyworm status impacts and control options in Africa (Evidence Note Update October–2018). CABI. Pp 1–53
Sharanabasappa D, Kalleshwaraswamy CM, Asokan R, MahadevaSwamy HM, Maruthi MS, Pavithra HB, Hedge K, Navi S, Prabhu ST, Goergen G (2018) First report of the fall armyworm, Spodoptera frugiperda (J E Smith) (Lepidoptera: Noctuidae), an alien invasive pest on maize in India. Pest Manag Hort Ecosyst 24:23–29
Shylesha AN, Jalali SK, Gupta A, Varshney R, Venkatesan T, Shetty P, Ojha R, Ganiger PC, Navik O, Subaharan K, Bakthavatsalam N, Ballal CR (2018) Studies on new invasive pest Spodoptera frugiperda (J. E. Smith) (Lepidoptera: Noctuidae) and its natural enemies. J Biol Control 32:145–151
Sisodiya DB, Raghunandan BL, Bhatt NA, Verma HS, Shewale CP, Timbadiya BG, Borad PK (2018) The fall armyworm, Spodoptera frugiperda (JE Smith) (Lepidoptera: Noctuidae); first report of new invasive pest in maize fields of Gujarat, India. J Entomol Zool 6:2089–2091
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tang RWK, Yau C, Ng WC (2010) Identification of stomatopod larvae (Crustacea: Stomatopoda) from Hong Kong waters using DNA barcodes. Mol Ecol Res 10:439–448
Todd EL, Poole RW (1980) Keys and illustrations for the armyworm moths of the noctuid genus Spodoptera Guenée from the Western Hemisphere. Ann Entomol Soc Am 73:722–738
Watts C, Dopheide A, Holdaway R, Davis C, Wood J, Thornburrow D et al (2019) DNA metabarcoding as a tool for invertebrate community monitoring: a case study comparison with conventional techniques. Austral Entomol 8:e1000417. https://doi.org/10.1111/aen.12384
Wyckhuys K, O’Neil R (2006) Population dynamics of Spodoptera frugiperda Smith (Lepidoptera: Noctuidae) and associated arthropod natural enemies in Honduran subsistence maize. Crop Prot 25:1180
Acknowledgements
We acknowledge the support of our donors who make action on Invasives possible, in particular, UK Aid (DFID) and the Directorate-General for International Cooperation (DGIS, Netherlands). CABI is an international intergovernmental organization, and we gratefully acknowledge the core financial support from our member countries (and lead agencies) including the United Kingdom (Department for International Development), China (Chinese Ministry of Agriculture), Australia (Australian Centre for International Agricultural Research), Canada (Agriculture and Agri-Food Canada), Netherlands (Directorate-General for International Cooperation), and Switzerland (Swiss Agency for Development and Cooperation). See http://www.cabi.org/about-cabi/who-we-work-with/key-donors/ for full details.
Author information
Authors and Affiliations
Contributions
SY and AR conceived and designed research. SY conducted experiments. SY, MM and KA analyzed data. SY, MM and NS wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval
This article does not contain any studies involving humans or animals (vertebrates).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yousaf, S., Rehman, A., Masood, M. et al. Occurrence and molecular identification of an invasive rice strain of fall armyworm Spodoptera frugiperda (Lepidoptera: Noctuidae) from Sindh, Pakistan, using mitochondrial cytochrome c oxidase I gene sequences. J Plant Dis Prot 129, 71–78 (2022). https://doi.org/10.1007/s41348-021-00548-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41348-021-00548-6