Abstract
Purpose
In this paper, we propose an open-source deep learning-based computer-aided diagnosis system for breast ultrasound images based on the Breast Imaging Reporting and Data System (BI-RADS).
Methods
Our dataset with 8,026 region-of-interest images preprocessed with ten times data augmentation. We compared the classification performance of VGG-16, ResNet-50, and DenseNet-121 and two ensemble methods integrated the single models.
Results
The ensemble model achieved the best performance, with 81.8% accuracy. Our results show that our model is performant enough to classify Category 2 and Category 4/5 lesions, and data augmentation can improve the classification performance of Category 3.
Conclusion
Our main contribution is to classify breast ultrasound lesions into BI-RADS assessment classes that place more emphasis on adhering to the BI-RADS medical suggestions including recommending routine follow-up tracing (Category 2), short-term follow-up tracing (Category 3) and biopsies (Category 4/5).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Breast cancer, the most common malignancy among women worldwide, has a high incidence rate among females aged 45–59 years. According to the World Health Organization’s 2020 statistics, 136.8 out of every 100,000 women in this age group were newly diagnosed with breast cancer, with 37.3 of them dying from it. This is three times higher than the second-highest incidence of colorectal cancer [1]. However, according to the American Cancer Society, patients with stage 0 and stage 1 breast cancer have a five-year survival rate of over 99% [2]. Therefore, promoting regular breast screening, especially among those at high-risk of breast cancer, can achieve the WHO’s breast cancer control strategy of early detection and early treatment [3]. The American College of Radiology (ACR) published the latest Breast Imaging Reporting and Data Systems (BI-RADS) 5th Edition [4] in 2013, which provides standard lesion descriptions, lesion risk evaluations, and medical action advice for mammography, sonography, and magnetic resonance imaging of the breast. Based on the pretest probability of pathological examination, BI-RADS assessment classifies lesions into seven categories that represent the integrative result of breast screening. Determining the false positive rate with an auxiliary screening tool is common in breast cancer screening, and the biopsy test is the necessary workup for breast cancer diagnosis.
It is noteworthy that Category 2 (with a 0% probability of malignancy) represents benign findings such as cysts, and routine follow-up tracing of the patient is recommended. Category 3 (with less than 2% probability of malignancy) represents probably benign findings such as fibroadenoma, and short-interval follow-up tracing is recommended. Category 4 and Category 5 (with 2–95% and > 95% probability of malignancy, respectively) represent suspicious and highly suggestive findings and are suggested to consider a biopsy test for further confirmation. The use of the BI-RADS standard lesion description can significantly reduce the variability of diagnosis reports, decrease intra-observer variation, and achieve intra-observer agreement. In addition, deep learning-based computer-aided diagnosis (CADx) systems are capable of not only reducing the time required for the task but also decreasing inter-observer variability through the use of an objective second opinion) [5,6,7,8]. Unlike machine learning-based CADx which has three stages (lesion detection, segmentation, and classification [5,6,7,8], deep learning-based CADx uses an end-to-end model to implement the lesion diagnosis task. This approach not only avoids the possibility of causing nonlesion options by using traditional image processing methods in the lesion detection stage but also decreases the dependence on the handcrafted segmentation feature [5].
The problems of gradient vanishing and degradation become more severe as convolutional neural networks (CNNs) become deeper. To address these issues, recent developments including ResNet [9, 10] and DenseNet [11] structures with skip connection and dense connection, respectively, promote forward propagation, and backpropagation and simultaneously improve the efficiency of models. Also, transfer learning is a way to deal with the rarity of medical images [12]. The study conducted by Brem et al. [13] highlights the importance of accurately classifying lesion risk levels and appropriately confirming them through pathological examinations during breast screening. Although the use of breast ultrasonography as an auxiliary diagnostic tool can increase the sensitivity of mammography on dense breasts from 47.6 to 76.1%, the false positive rate could remain as high as 2.4%.
In this paper, our main contribution is to establish a deep learning-based CADx system for classifying breast ultrasound lesions into three categories, in which patients require routine follow-up tracing (Category 2), short-term follow-up tracing (Category 3), and biopsies (Category 4/5).
2 Literature Review
2.1 The Diagnosis of Breast Lesions
Mammography, sonography, and magnetic resonance imaging (MRI) are three common imaging methods for breast screening. Mammography, which uses low radiation doses, is the most widely used method for first-line screening. Sonography is often used to detect obscured lesions, especially in dense breasts. MRI is more commonly used to screen high-risk patients due to its high cost and time-consuming nature [6, 14]. Computer-aided systems for breast lesion analysis can be classified into two types: computer-aided detection (CADe) systems, which are used to detect the position of lesions; and the CADx systems, which aid decision-making for lesion classification. The structure of a CADx system for breast lesion analysis typically includes three steps: lesion detection, lesion segmentation, and lesion classification [5, 6]. Deep learning-CADx systems, which differ from traditional machine learning-based methods, reduce the dependence on handcrafted features and decrease human intervention (and the bias that comes with it) through automatic feature extraction [5, 8, 15].
Since mammography enables the visualization of the entire breast architecture, studies of deep learning-based CADx systems typically involve two stages: lesion localization and malignant classification. Ribli et al. [16] utilized a Fast R-CNN model with a Region Proposal Network for lesion detection and Fully Connected Layers for classification, achieving an area under the curve (AUC) of 0.95 and 90% sensitivity for malignant lesions. Al-Antari et al. [17] used the YOLO model for lesion detection, and ResNet-50, and InceptionResNet-v2 for classification, achieving accuracy rates of 92.55% and 95.32%, respectively. It is noteworthy that all of these studies were conducted on the INBreast dataset, which consists of digital mammography images; thus, the performances of these models on other datasets and modalities may vary. As for calcified lesions, Rehman et al. [18] not only automatically segmented the region-of-interest (ROI) of lesions but also utilized the depthwise-separable convolutional layers to classify them into benign and malignant lesions. The performance achieved 80% and 87% accuracy before and after model improvement, respectively.
Conversely, there are some previous studies in which the breast ultrasound CADx system is used in the diagnosis of benign and malignant lesions. Han et al. [19] utilized a single model structure, GoogleNet, and achieved 91.2% mean accuracy. Moreover, they also found the best ROI selection method was the distance between the lesion’s margin and the ROI’s edge, with 180 pixels being the optimal threshold. Qi et al. [20] focused on the double labeling of malignant and solid lesions. They performed cross-training of the Inception-ResNet-v2 and Inception-v3 models, which resulted in 94.5% accuracy. Besides, Moon et al. [21] utilized fusion imaging and the ensemble model with average prediction to achieve the best accuracy rate (91.1%). These previous studies demonstrate the potential of breast ultrasound CADx systems in accurately diagnosing breast lesions. Moreover, there is a growing demand for breast MRI CADx systems to deal with 3D imaging for more in-depth analyses of breast tissues in clinical settings. Balkenende et al. investigated various methods of breast MRI CADx systems and highlighted 3D image usage and extraction [14]. In one study, Zhou et al. [22] used a single 2D MRI image as a unit and extracted the ROI via morphological methods. Then, they used an ensemble model consisting of two types of DenseNet to distinguish benign breast lesions from malignant ones, achieving 83.7% accuracy [22].
2.2 Assessment of Ultrasound Images
A thorough review of several studies revealed that the application of machine learning and deep learning in the sonography CADx system is not only limited to breast lesions but is also utilized in diagnosing lesions in other organs [15]. For BI-RADS risk level diagnosis in breast sonography, Huang et al. [23] utilized a two-stage model of the fully convolutional network and the deep CNN to detect lesions and sequentially distinguish lesions’ risks. They achieved 99%, 94%, 73%, 92%, and 87% accuracy for BI-RADS categories 3, 4 A, 4B, 4 C, and 5, respectively. Moreover, Ciritsis et al. [24] used the sliding window method and four different deep CNNs to hierarchically implement automatic breast lesion classification, obtaining an AUC of 0.84.
Chang et al. [25] used the Speckle Reducing Anisotropic Diffusion and the K-means clustering methods to segment lesions and extract features. They used three classifiers of the SVM, the Random Forest, and the CNN to distinguish BI-RADS Category 2–5, obtaining accuracy rates of 80%, 77.8%, and 85.4%, respectively. Furthermore, Xing et al. [26] provided the BI-RADS Vector-Attention Network, which used the Residual Attention Block to extract features and incorporate the assemble vector of the BI-RADs standard description to distinguish between benign lesions and malignant ones. Similarly, Qian et al. [27] utilized a combined fusion image from three different ultrasound modalities to train the Squeeze-and-Excitation Network for lesion malignancy classification. They also used the transverse and longitudinal views to comprehensively evaluate lesion malignancy and effectively decreased the false-positivity rate of single-view evaluation. Finally, they achieved an AUC of 0.96 [95% CI: 0.91–0.98]. These reviewed studies demonstrate that applying deep learning methods to implement ultrasound CADx systems is effective and combining different models’ opinions by the ensemble methods can enhance classification efficiency [20, 22, 27].
3 Materials and Methods
3.1 Dataset
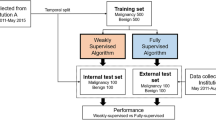
Our study was approved by the National Yang Ming Chiao Tung University’s Institutional Review Boards under project number YM108127E. All the ultrasound images used in this paper were captured using an ultrasound device (Terason uSmart 3300, Probe Medical Corporation) with a depth of 4 cm. Each image in the dataset was carefully labeled by a professional radiologist and a physician trained in breast surgery. The dataset contains 569 cases, with a total of 8,026 ROI images. These images are further categorized as follows: 3,662 images belonged to Category 2 (lesion size: 0.77 ± 0.26 cm), 2,390 images belonged to Category 3 (lesion size: 1.23 ± 0.45 cm), and 1,974 images belonged to Category 4/5 (lesion size: 1.90 ± 0.65 cm).
To prepare the dataset for training and testing purposes, it was divided into two, with 80% being allocated to the training set and 20% to the test set. Before feeding the images into the model, they were preprocessed with histogram equalization, and their pixel values were rescaled to 0–1 for normalization. Also, we employed one-hot encoding to represent the labeling data. To increase the diversity of the dataset and improve the model’s performance, we augmented the images by applying horizontal flipping and translation with a magnitude of 12 pixels ten times, which resulted in an expanded dataset. Finally, all images were resized to a resolution of 128 × 128 pixels, ensuring dataset uniformity.
3.2 Deep Learning Models
A server equipped with a graphics processing unit (NVIDIA GeForce GTX 1080 Ti, NVIDIA Corporation) was used for experiments in our study. We also utilized Tensorflow to build the deep learning framework and Tkinter to design our CADx interface. Our study encompassed three types of deep learning architecture: VGG [28], ResNet [10], and DenseNet [11]. These different types of architecture were initialized with ImageNet pretrained models and fine-tuned with breast ultrasound ROI images. During the training process, we employed a mini-batch size of 32 images and utilized Stochastic Gradient Descent with a learning rate of \({10}^{-5}\)and a momentum of 0.9 as an optimizer. Additionally, to mitigate model overfitting, a weight decay of \({10}^{-5}\) and a dropout rate of 0.5 were incorporated. For the analysis of categorical results, we used several evaluation metrics, including precision, recall, F1-score, and AUC [29]. Furthermore, the accuracy and the balanced accuracy [30] were also used to evaluate the model’s performance.
3.3 Ensemble Methods
Herein, we employed three different types of deep learning architecture that showed the best performance to build our ensemble model. To combine their predictions, we utilized two ensemble methods, which are average prediction and voting. For the average prediction method, we calculated the average of the three models’ predictions for each set of test data. The category with the highest average was selected as the final predicted category. For the voting method, we calculated the number of predicted categories from the three models. The category with the highest number of votes was chosen as the final prediction of the ensemble model.
In cases where the first and second models had different opinions on the predicted category, the third model’s vote played a crucial role in determining the final prediction. However, if these three single models had completely distinct opinions, indicating significant disagreement, we chose the medium risk level (BI-RADS Category 3) as the final predicted category of the ensemble model. This decision aimed to mitigate extreme predictions and provide a more conservative assessment in cases of conflicting model outputs.
3.4 Structure of Our Deep Learning-Based Computer-Aided Diagnosis (CADx) System
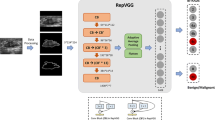
Based on breast ultrasound raw data and deep learning techniques, we propose a comprehensive CADx system for automated diagnosis and human interpretation. By utilizing the BI-RADS standard description, this system can be used to not only improve case follow-up but also optimize the deep learning performance of our breast ultrasound CADx system, which can be seamlessly integrated with a picture archiving and communication system (PACS) and can also be applied in a routine imaging workflow. This integrated system provides two main functions: a labeling breast ultrasound lesion service and a retrieving and deleting patient report service. Additionally, our system goes beyond the basic functionalities and focuses on the analysis and management of the diagnosis report database, including a statistical analysis service and a missing data check service. To address patients’ privacy concerns while optimizing deep learning performance, our system provides a de-identification service that ensures the appropriate protection of patient data, adhering to privacy regulations. The system architecture is illustrated in Fig. 1.
4 Results and Discussion
4.1 The Evaluation of the Single Models
Table 1. presents the results of different single models used in our study. Among these models, the RestNet-50 model achieved the best performance with 80.7% accuracy and 79.2% balanced accuracy with histogram equalization and data augmentation. It is noteworthy that correctly classifying BI-RADS Category 3, which represents the medium risk level of lesions, posed a challenge in every single model’s results. The precision, recall rate, and F1 score of this category were relatively lower than those of the other categories. This indicates the difficulty in accurately distinguishing lesions in the medium risk category. These performance results provide valuable insights into the strengths and limitations of the single models, highlighting the challenges associated with the correct classification of lesions in the BI-RADS Category 3.
In our study, we compared the results of the VGG-16, ResNet-50, and DenseNet-121 models, and analyzed their Grad-CAM heat maps. Despite the complexity and rarity of the breast ultrasound dataset, we observed interesting variations in the heat maps generated by different models, even when they made the same predictions for a given example. The Grad-CAM [31] heat map results are presented in Fig. 2.
Heat maps of different single models. (A), (E), and (I) are the ROI examples of the BI-RADS Category 2, Category 3, and Category 4/5 lesions, respectively. (B), (F), and (J) are the visualization results of Grad-CAM using the VGG model. (C), (G), and (K) are the visualization results of Grad-CAM using the ResNet model. (D), (H), and (L) are the visualization results of Grad-CAM using the DenseNet model
We found that the ResNet-50 and DenseNet-121 models produced more precise and detailed heat maps than the VGG-16 model produced, indicating that these models were able to capture and highlight important regions within the breast ultrasound images with higher accuracy and precision. For example, in Fig. 2(C), the ResNet-50 model accurately detected and emphasized the enhancement of the ultrasound posterior feature in a Category 2 lesion example. This demonstrates the model’s ability to capture subtle and important lesion characteristics. Furthermore, the Grad-CAM heat maps provided insights into the challenges associated with the classification of BI-RADS Category 3 lesions. The variations in heat maps among different models indicate the difficulty in accurately distinguishing and localizing features of lesions in this category.
4.2 The Evaluation of the Ensemble Model
We applied ensemble methods (average prediction and voting) using the best-performing VGG-16, ResNet-50, and DenseNet-121 models trained with histogram equalization and data augmentation. The results of the ensemble models are presented in Table 2. We observed that the ensemble methods not only improved the overall performance of the model but also increased the classification efficiency for the challenging BI-RADS Category 3. They showed comparable performances to those of the average prediction and voting methods. However, the ensemble model with average prediction consistently achieved better AUC values compared to the voting method. This can be attributed to the fact that the voting method reduces the sensitivity of the predicted numbers, affecting the discernment. To further illustrate the performance, the receiver operating characteristic (ROC) curves of the ensemble methods with average prediction and voting are presented in Fig. 3, and Fig. 4, respectively.
4.3 Comparisons with a State-of-the-Art Study
We compared the results of our study to Hejduk’s method [32], and the comparison is presented in Table 3. Our study achieved an accuracy rate of 79.7% and 77.6% balanced accuracy, outperforming Hejduk et al.’s study regarding both the best performance of the single models and the best performance of the ensemble models. It is noteworthy that our study differed from Hejduk’s method regarding the classification approach. While Hejduk’s study focused on binary risk classes of BI-RADS Category 2/3 and BI-RADS Category 4/5, ours emphasized adhering to the BI-RADS medical suggestions. This included recommending routine follow-up for BI-RADS Category 2, short-term follow-up for BI-RADS Category 3, and a biopsy for BI-RADS Category 4/5. This alignment with practical application scenarios adds relevance and usability to our study’s findings.
4.4 Implementation of Our Deep Learning-Based Computer-Aided Diagnosis System (CADx)
To promote the development of computer-aided diagnosis systems, we are also dedicated to opening our computer-aided diagnosis system for use in research. Our CADx system implemented communication interfaces regarding of DICOM DIMSE-C Services to ensure seamless integration with PACS. The deidentification module utilizes the guidelines provided by the DICOM Basic Attribute Confidentiality Profiles [33] to deidentify breast ultrasound DICOM objects and derivative patient reports, maintaining attribute confidentiality, and privacy throughout the diagnostic workflow .As shown in Fig. 5, the main page of our CADx system concludes five different areas: (A) the Setting Box, (B) DICOM’s Pixel Data Display Area, (C) the Control Box, (D) the Patient Report Display Area, and (E) Other Areas. The detailed source code of our proposed CADx and execution proposed AI model are public available on the GitHub [34].
In the routine workflow of breast ultrasound image screening, sonographers or physicians conduct real-time ultrasound scanning, selecting key images that are subsequently uploaded to PACS. Following this, physicians retrieve these key images from PACS, reviewing and interpreting them for reporting purposes. The proposed CADx system assists physicians in interpreting images and generating standardized reports, aiding in classifying BI-RADS assessment classes and facilitating data statistical analysis and reuse through standardized data fields that prioritize adherence to BI-RADS medical guidelines. This improvement can facilitate physicians in reporting with the classification of BI-RADS. However, this scenario does not realize real-time assessment during screening.
To integrate this AI model, separating from the CADx system, into real-time ultrasound scanning in clinics, it could be combined with ultrasound devices equipped with higher-speed computing units to enable real-time detection during examinations. Additionally, deploying the CADx as a cloud service or a cloud PACS with high network bandwidth to swiftly retrieve the AI results could be another suggestion. Furthermore, enhancing performance by integrating image acquisition information such as echo patterns and posterior features through the concept of multi-modality into this AI model integrated with ultrasound devices could further enhance the interpretation of BI-RADS features in the future.
5 Conclusions
Our study contributes significantly to the field of breast ultrasound lesion classification by employing deep learning models. We achieved competitive performance in accurately classifying breast ultrasound lesions into the ACR BI-RADS assessment of Category 2, Category 3, and Category 4/5. We also developed an open-source computer-aided diagnosis system to facilitate the practical application of our research. Our study focused on achieving accurate and reliable classification results, which are crucial for determining the risk level of breast lesions and providing appropriate medical recommendations. We employed various deep learning models, with ResNet-50 yielding the best performance with 80.7% accuracy and 79.2% balanced accuracy. Furthermore, we proposed the ensemble models and found that the average prediction approach further improved the classification performance with 81.8% accuracy and 80.2% balanced accuracy.
Our results are comparable to Hejduk’s method, which signifies the quality and competitiveness of our findings in the context of breast lesion classification. Also, we found that training models with histogram equalization and data augmentation led to notable improvements in accuracy (3–5%). Additionally, the utilization of data augmentation and ensemble methods enhanced the performances of all models in classifying BI-RADS Category 3 lesions. Our experimental results demonstrated excellent classification performances (AUCs > 0.9) for both BI-RADS Category 2 and Category 4 and good performance (0.8 < AUC < 0.9) for BI-RADS Category 3, indicating the reliability and efficacy of our findings in differentiating lesion categories.
To further improve our study, we recommend expanding the dataset to encompass a broader range of ROIs, incorporating additional ACR BI-RADS standard lesion descriptions such as shape, margin, and echo pattern features for improved classification, and standardizing the ROI by quantifying the surrounding tissues of lesions [19].
Data Availability
The experimental datasets used in this manuscript is unavailable due to privacy or ethical restrictions.
References
Ferlay, J., Colombet, M., Soerjomataram, I., Parkin, D. M., Piñeros, M., Znaor, A., & Bray, F. (2021). Cancer statistics for the year 2020: An overview. International Journal of Cancer, 149(4), 778–789. https://doi.org/10.1002/ijc.33588.
American Cancer Society (2023, March 9). Cancer Statistics Center. Retrieved March 9, 2024, from http://cancerstatisticscenter.cancer.org.
Ginsburg, O., et al. (2020). Breast cancer early detection: A phased approach to implementation. Cancer, 126(S10), 2379–2393. https://doi.org/10.1002/cncr.32887.
American College of Radiology (2013, March 9). ACR BI-RADS Atlas: breast imaging reporting and data system, 5th ed., Reston, VA: American College of Radiology. Retrieved March 9, 2024, from https://www.acr.org/Clinical-Resources/Reporting-and-Data-Systems/Bi-Rads.
Lu, L., Zheng, Y., Carneiro, G., & Yang, L. (2017). Deep learning and convolutional neural networks for medical image computing. Springer.
Hassan, N. M., Hamad, S., & Mahar, K. (2022). Mammogram breast cancer CAD systems for mass detection and classification: A review. Multimedia Tools and Applications, 81(14), 20043–20075. https://doi.org/10.1007/s11042-022-12332-1.
Bagchi, S., Tay, K. G., Huong, A. K. C., & Debnath, S. K. (2020). Image processing and machine learning techniques used in computer-aided detection system for mammogram screening - a review. International Journal of Electrical and Computer Engineering, 10(3), 2336–2348. https://doi.org/10.11591/ijece.v10i3.pp2336-2348.
Loizidou, K., Elia, R., & Pitris, C. (2023). Computer-aided breast cancer detection and classification in mammography: A comprehensive review. Computers in Biology and Medicine, 153, 106554. https://doi.org/10.1016/j.compbiomed.2023.106554.
He, K., Zhang, X., Ren, S., & Sun, J. (2016). Identity mappings in deep residual networks. Proceeding of ECCV2016, 630-645. https://doi.org/10.1007/978-3-319-46493-0_38.
He, K., Zhang, X., Ren, S., & Sun, J. (2016). Deep residual learning for image recognition. Proceeding of CVPR 2016, 770-778. https://doi.org/10.1109/CVPR.2016.90.
HuangG., LiuZ., MaatenL. V. D., & WeinbergerK. Q. (2017). Densely connected convolutional networks. Proceeding of CVPR 2017, 2261-2269. https://doi.org/10.1109/CVPR.2017.243.
Tan, C., Sun, F., Kong, T., Zhang, W., Yang, C., & Liu, C. (2018). A survey on deep transfer learning. Proceeding of International Conference on Artificial Neural Networks 2018, 270–279. Available https://link.springer.com/chapter/https://doi.org/10.1007/978-3-030-01424-7_27.
Brem, R. F., Lenihan, M. J., Lieberman, J., & Torrente, J. (2015). Screening breast ultrasound: Past, present, and future. American Journal of Roentgenology, 204(2), 234–240. https://doi.org/10.2214/AJR.13.12072.
Balkenende, L., Teuwen, J., & Mann, R. M. (2022). Application of deep learning in breast Cancer imaging. Seminars in Nuclear Medicine, 52(5), 584–596. https://doi.org/10.1053/j.semnuclmed.2022.02.003.
Liu, S., Wang, Y., Yang, X., Lei, B., Liu, L., Li, S., Ni, D., & Wang, T. (2019). Deep learning in medical ultrasound analysis: A review. Engineering, 5(2), 261–275. https://doi.org/10.1016/j.eng.2018.11.020.
Ribli, D., Horváth, A., Unger, Z., Pollner, P., & Csabai, I. (2018). Detecting and classifying lesions in mammograms with deep learning. Scientific Reports, 8(1), 4165. https://doi.org/10.1038/s41598-018-22437-z.
Al-antari, M. A., Han, S. M., & Kim, T. S. (2020). Evaluation of deep learning detection and classification towards computer-aided diagnosis of breast lesions in digital X-ray mammograms. Computer Methods and Programs in Biomedicine, 196, 105584. https://doi.org/10.1016/j.cmpb.2020.105584.
Rehman, K. U., Li, J., Pei, Y., Yasin, A., Ali, S., & Mahmood, T. (2021). Computer vision-based microcalcification detection in digital mammograms using fully connected depthwise sepadrable convolutional neural network. Sensors (Basel, Switzerland), 21(14), 4854. https://doi.org/10.3390/s21144854.
Han, S., Kang, H. K., Jeong, J. Y., Park, M. H., Kim, W., Bang, W. C., & Seong, Y. K. (2017). A deep learning framework for supporting the classification of breast lesions in ultrasound images. Physics in Medicine & Biology, 62(19), 7714–7728. https://doi.org/10.1088/1361-6560/aa82ec.
Qi, X., Zhang, L., Chen, Y., Pi, Y., Chen, Y., Lv, Q., & Yi, Z. (2019). Automated diagnosis of breast ultrasonography images using deep neural networks. Medical Image Analysis, 52, 185–198. https://doi.org/10.1016/j.media.2018.12.006.
Moon, W. K., Lee, Y. W., Ke, H. H., Lee, S. H., Huang, C. S., & Chang, R. F. (2020). Computer-aided diagnosis of breast ultrasound images using ensemble learning from convolutional neural networks. Computer Methods and Programs in Biomedicine, 190, 105361. https://doi.org/10.1016/j.cmpb.2020.105361.
Zhou, J., Luo, L. Y., Dou, Q., Chen, H., Chen, C., Li, G. J., Jiang, Z. F., & Heng, P. A. (2019). Weakly supervised 3D deep learning for breast cancer classification and localization of the lesions in MR images. Journal of Magnetic Resonance Imaging, 50(4), 1144–1151. https://doi.org/10.1002/jmri.26721.
Huang, Y., Han, L., Dou, H., Luo, H., Yuan, Z., Liu, Q., Zhang, J., & Yin, G. (2019). Two-stage CNNs for computerized BI-RADS categorization in breast ultrasound images. BioMedical Engineering OnLine, 18(1), 8. https://doi.org/10.1186/s12938-019-0626-5.
Ciritsis, A., Rossi, C., Eberhard, M., Marcon, M., Becker, A. S., & Boss, A. (2019). Automatic classification of ultrasound breast lesions using a deep convolutional neural network mimicking human decision-making. European Radiology, 29(10), 5458–5468. https://doi.org/10.1007/s00330-019-06118-7.
Chang, Y. W., Chen, Y. R., Ko, C. C., Lin, W. Y., & Lin, K. P. (2020). A Novel Computer-aided-diagnosis system for breast ultrasound images based on BI-RADS categories. Applied Sciences, 10(5), 1830. https://doi.org/10.3390/app10051830.
Xing, J., Chen, C., Lu, Q., Cai, X., Yu, A., Xu, Y., Xia, X., Sun, Y., Xiao, J., & Huang, L. (2021). Using BI-RADS stratifications as auxiliary information for breast masses classification in ultrasound images. IEEE Journal of Biomedical and Health Informatics, 25(6), 2058–2070. https://doi.org/10.1109/jbhi.2020.3034804.
Qian, X., Pei, J., Zheng, H., Xie, X., Yan, L., Zhang, H., Han, C., Gao, X., Zhang, H., Zheng, W., Sun, Q., Lu, L., & Shung, K. K. (2021). Prospective assessment of breast cancer risk from multimodal multiview ultrasound images via clinically applicable deep learning. Nature Biomedical Engineering, 5(6), 522–532. https://doi.org/10.1038/s41551-021-00711-2.
Liu, S., & Deng, W. (2015). Very deep convolutional neural network based image classification using small training sample size. Proceeding of IEEE ACPR 2015, 730–734. https://doi.org/10.1109/ACPR.2015.7486599.
Nahm, F. S. (2022). Receiver operating characteristic curve: Overview and practical use for clinicians. Korean Journal of Anesthesiology, 72(1), 25–36. https://doi.org/10.4097/kja.21209.
Grandini, M., Bagli, E., & Visani, G. (2020). Metrics for Multi-Class Classification: An overview, arXiv. Available https://doi.org/10.48550/arXiv.2008.05756.
Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R., Parikh, D., & Batra, D. (2017). Grad-CAM: Visual explanations from deep networks via gradient-based localization. IEEE ICCV, 2017, 618–626. https://doi.org/10.1109/ICCV.2017.74.
Hejduk, P., Marcon, M., Unkelbach, J., Ciritsis, A., Rossi, C., Borkowski, K., & Boss, A. (2022). Fully automatic classification of automated breast ultrasound (ABUS) imaging according to BI-RADS using a deep convolutional neural network. European Radiology, 32(7), 4868–4878. https://doi.org/10.1007/s00330-022-08558-0.
DICOM Supplement 142 Clinical Trial De-identification Profiles, NEMA Standard (2024, March 9). Retrieved March 9, 2024, from https://dicom.nema.org/medical/dicom/Final/sup142_ft.doc.
Github, U. I. (2024, March 9). Retrieved March 9, 2024, from https://github.com/SuHsinYa/Computer-Aided-Diagnosis-System-for-Breast-Ultrasound-Images/tree/master.
Acknowledgements
This research was funded by National Science and Technology Council, Taiwan, grant number 112-2634-F-006-002 and 112-2221-E-227-001. We appreciate the New England Clinic, Taipei, Taiwan, and the effort of Dr. Huang and Xiao radiologist in data labeling.
Funding
This research was funded by National Science and Technology Council, Taiwan, grant number 112-2634-F-006-002 and 112-2221-E-227-001.
Open Access funding enabled and organized by National Yang Ming Chiao Tung University
Author information
Authors and Affiliations
Contributions
All authors contributed to the study’s conception and design. Material preparation and data collection were performed by Hsin-Ya Su and Pai-Jung Huang. Data analysis and validation were performed by Hsin-Ya Su and Chung-Yueh Lien. The software was developed by Hsin-Ya Su and Chung-Yueh Lien. The first draft of the manuscript was written by Hsin-Ya Su, Chung-Yueh Lien, and Woei-Chyn Chu. Review and editing were written by Chung-Yueh Lien and Woei-Chyn Chu. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of National Yang Ming Chiao Tung University (protocol code YM108127E and date of approval).
Consent to Participate
Patient consent was waived due to the retrospective nature of the study and the use of deidentified clinical data.
Consent to Publish
Patient consent was waived due to the personal identifiers in Figs. 2 and 5 being complete de-identified.
Competing Interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Su, HY., Lien, CY., Huang, PJ. et al. A Practical Computer Aided Diagnosis System for Breast Ultrasound Classifying Lesions into the ACR BI-RADS Assessment. J. Med. Biol. Eng. 44, 426–436 (2024). https://doi.org/10.1007/s40846-024-00869-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40846-024-00869-5