Abstract
Introduction
Research has highlighted the role of runt-related transcription factor 2 (Runx2) in the development of osteoarthritis (OA); however, its causal association remains unclear. This study aimed to explore whether Runx2 expression is causally associated with OA and assess its therapeutic potential for OA.
Methods
Genetic proxy instruments for Runx2 expression were obtained from gene expression quantitative trait locus (eQTLs) study of eQTLGen Consortium (n = 31,684). Aggregated genome-wide association study (GWAS) data for OA (including all OA [177,517 cases and 649,173 controls], knee OA (KOA) [62,497 cases and 333,557 controls], and hip OA (HOA) [36,445 cases and 316,943 controls]) were extracted from the Genetics of Osteoarthritis Consortium. We integrated eQTLs data with OA GWAS data to estimate their causal association and to estimate the potential of Runx2 as a drug target in the treatment of OA using summary data-based Mendelian randomization (SMR) analysis. Furthermore, different OA GWAS data (including all OA [77,052 cases and 378,169 controls], KOA [24,955 cases and 378,169 controls], and HOA [15,704 cases and 378,169 controls]) derived from the GWAS Catalog database were used for replication study.
Results
SMR analysis showed that high expression levels of Runx2 were associated with an increased risk of all OA [odds ratio (OR) 1.044, 95% confidence interval (CI) 1.023–1.067; P = 5.03 × 10−5], KOA (OR 1.040, 95% CI 1.006–1.075; P = 0.021), and HOA (OR 1.067, 95% CI 1.022–1.113; P = 0.003). This suggests that Runx2 inhibitors may have promising potential for the treatment of OA. Notably, the causal effects of Runx2 with all OA (OR 1.053, 95% CI 1.027–1.079; P = 3.95 × 10−5) and KOA (OR 1.043, 95% CI 1.001–1.087; P = 0.045) were repeated in the replication study, but limited evidence supported the association of Runx2 expression levels with HOA (OR 1.045, 95% CI 0.993–1.101; P = 0.094).
Conclusions
Our analyses indicate a positive correlation between Runx2 expression and OA risk across all three phenotypes, suggesting the potential of Runx2 inhibitors in the treatment of OA and providing evidence from a genetic perspective.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Why carry out this study? |
Research has highlighted the role of runt-related transcription factor 2 (Runx2) in the development of osteoarthritis (OA); however, its causal association remains unclear. |
This study aimed to explore whether Runx2 expression is causally associated with OA and assess its therapeutic potential for OA using summary data-based Mendelian randomization (SMR) analysis. |
What was learned from the study? |
Expression levels of Runx2 were positively associated with the risk of OA, implying that Runx2 inhibitors have great potential for the treatment of OA. |
Our study provides further support for the development of drugs targeting Runx2 for the treatment of OA from a genetic perspective, providing new insights into the treatment strategies for OA. |
Introduction
Osteoarthritis (OA) is a degenerative disease caused by various factors that lead to fibrosis, cracking, ulceration, and loss of articular cartilage, with joint pain being the main symptom [1]. It can involve multiple joint sites, particularly the knees, hips, and hands [2]. The prevalence of OA is extremely high, affecting approximately 500 million people worldwide [3]. As populations age, its prevalence has risen. Statistically, by the middle of the twenty-first century, over one billion people are anticipated to be affected by this disease worldwide [4], which will bring an enormous burden to society and patients. However, existing treatments fail to meet satisfactory expectations [5]. Arthroplasty is the only established and effective method for treating patients with end-stage OA [6]. Therefore, the exploration of new drug targets for OA is urgently needed.
Runt-related transcription factor 2 (Runx2), a transcription factor, is one of the critical members of the Runx family. Runx2 plays an important regulatory role in the process of chondrocyte maturation and osteoblast osteogenesis [7, 8]. Accumulating evidence has shown that Runx2 contributes to the progression of OA [2, 9]. Several studies have observed the upregulation of Runx2 in both human and animal OA cartilage samples [9, 10]. Catheline et al. demonstrated that specific overexpression of Runx2 in cartilage exacerbated cartilage destruction in traumatic OA using a surgical model of meniscus/ligament injury [11]. Additionally, a promising study revealed that Runx2 synergizes with distal-less homeobox 5 to promote the expression of type X collagen (Col10a1) and chondrocyte hypertrophy, which in turn leads to the development of OA [12]. Notably, after the specific knockdown of Runx2 in the chondrocytes of mice undergoing destabilization of the medial meniscus, the mice exhibited a lighter OA phenotype [13]. Moreover, Huang et al. found that the intra-articular delivery of an adeno-associated virus to a mouse OA model significantly decreased Runx2 expression by increasing miR-204 expression, thereby alleviating the symptoms and pain of OA in mice [14]. Evidently, Runx2 is a promising therapeutic target for OA. Unfortunately, to date, no drugs targeting Runx2 for the treatment of OA have been developed.
Mendelian randomization (MR) is a novel method that uses genetic variation as instrumental variable (IV) to infer causal association between exposures and outcomes. This effectively eliminates the interference of confounding factors and reverse causation, resulting in greatly improved accuracy and credibility of the results [15]. As large-scale genome-wide association studies (GWAS) and expression quantitative trait loci (eQTL) continue to be explored, we have the opportunity to integrate them to explore susceptibility risk genes for different diseases [16]. Moreover, as MR provides novel insights into the pathogenesis of diseases at the genetic level, it is often used to develop drug targets [17]. For example, MR has been successfully used to identify potential therapeutic targets for atrial fibrillation, major depressive disorder, and multiple cancers [18,19,20].
To date, no MR studies have been performed to identify a causal association between Runx2 gene expression and OA. Therefore, this study aimed to explore the causal relation between the expression levels of Runx2 and OA (including all OA, knee OA [KOA], and hip OA [HOA]) and to estimate its potential in the treatment of OA using summary data-based MR (SMR) analysis.
Methods
Study Design
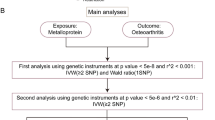
Figure 1 illustrates the study design and workflow. In current study, we used blood expression levels of Runx2 from eQTLs as exposure and OA-aggregated GWAS data (including all OA, KOA, and HOA) as outcomes to investigate their causal association and estimate the potential of Runx2 as a drug target for OA. The SMR analysis was used as the primary method. Additionally, to improve the reliability of the results, we performed a heterogeneity in dependent instrument (HEIDI) test. Summarized OA GWAS data from the Genetics of Osteoarthritis (GO) Consortium were used as the primary discovery study (https://msk.hugeamp.org/downloads.html), and OA GWAS data from the UK Biobank and Arthritis Research UK Osteoarthritis Genetics (arcOGEN) Consortium were used as the replication study [21].
Data Sources
The eQTLs data for blood expression of Runx2 were sourced from the eQTLGen Consortium, which identified 16,989 cis-eQTL genes in 31,684 blood samples [22]. In present analysis, only cis-eQTLs that reached the P < 5 × 10−8 threshold and were located within 1 Mb of the gene center were included (www.eqtlgen.org/cis-eqtls.html). Notably, genetic variants associated with gene expression in mtDNA and X and Y chromosomes were excluded [22]. In our discovery study, aggregated GWAS data for OA were sourced from the largest GWAS meta-analysis of OA, comprising 13 international cohorts and 826,690 individuals [23]. This study analyzed 11 OA phenotypes, of which three (all OA, KOA, and HOA) were used in our study. The summary data for all OA included 826,690 individuals (177,517 cases and 649,173 controls), the summary data for KOA included 396,054 individuals (62,497 cases and 333,557 controls), and the summary data for HOA included 353,388 individuals (36,445 cases and 316,943 controls) [23].
To validate our findings, we used different OA GWAS data provided by the UK Biobank and arcOGEN Consortium for the replication study (all OA, 77,052 cases and 378,169 controls; KOA, 24,955 cases and 378,169 controls; HOA, 15,704 cases and 378,169 controls) [21]. Additional data are presented in Table 1 and Supplementary Table 1.
Statistical Analysis
As an extension of traditional MR analysis, the SMR method facilitates the inference of pleiotropic associations between diverse molecular QTL data (e.g., eQTLs data) and diseases of interest [16]. Compared to traditional MR analyses, SMR using the top cis-QTL as the IV can yield higher statistical power when exposure and outcome are obtained from two independent samples with large sample sizes [16]. In the current SMR analysis, we checked the consistency of the allele frequency of each single-nucleotide polymorphism (SNP) between pairwise datasets, including eQTL, GWAS, and linkage disequilibrium reference data. SNPs with allele frequency differences < 0.20 between any pair of datasets were included. In addition, to minimize the bias caused by pleiotropy, the HEIDI test was employed. Of note, a P value of HEIDI > 0.01 suggested no pleiotropy, implying that the result was reliable [24]. Both SMR and HEIDI tests were performed using SMR software for Windows version 1.3.1.
Ethical Approval
Data used in our study were publicly available, each original study was approved by the respective institutional ethics review board, and informed consent was obtained from the participants. Therefore, further ethical approval was not required.
Results
Runx2 Gene Expression and OA
We identified 2293 cis-eQTLs (P < 5 × 10−8) for Runx2 expression from the eQTL consortium. According to the SMR principle, only the top cis-eQTL rs1200428 (most significant) was used as the IV to proxy the expression levels of Runx2. As shown in Fig. 2 and Supplementary Table 2, our SMR analysis found that each increase of one standard deviation in Runx2 expression levels elevated the risk of all OA by 4.4% (OR 1.044, 95% CI 1.023–1.067; P = 5.03 × 10−5), the risk of KOA by 4% (OR 1.040, 95% CI 1.006–1.075; P = 0.021), and the risk of HOA by 6.7% (OR 1.067, 95% CI 1.022–1.113; P = 0.003), respectively. Fortunately, the association of Runx2 with all OA (p_HEIDI test = 0.070), KOA (p_HEIDI test = 0.494), and HOA (p_HEIDI test = 0.020) was not driven by pleiotropy (Fig. 2, Supplementary Table 2), suggesting that the results were robust.
Results of the Replication Study
Identical to the discovery study, a total of 2293 cis-eQTLs (P < 5 × 10−8) associated with Runx2 expression were identified (Supplementary Table 2). Most significant top cis-eQTL (rs1200428) was applied to proxy the expression levels of Runx2. Our analysis showed that Runx2 expression levels were associated with an increased risk of all OA (OR 1.053, 95% CI 1.027–1.079; P = 3.95 × 10−5) and KOA (OR 1.043, 95% CI 1.001–1.087; P = 0.045), but not with the HOA risk (OR 1.045, 95% CI 0.993–1.101; P = 0.094) (Fig. 2, Supplementary Table 2). The HEIDI test suggested that the association of Runx2 with all OA (p_HEIDI test = 0.124), KOA (p_HEIDI test = 0.730), and HOA (p_HEIDI test = 0.201) was not affected by pleiotropy (Fig. 2, Supplementary Table 2).
Discussion
Using blood eQTLs data and GWAS summary data, we performed SMR analysis to explore the causal relation between Runx2 expression and OA risk and its potential as a drug target for OA. The results of our discover studies showed that high expression levels of Runx2 were associated with an increased risk of all three OA phenotypes (all OA, KOA, and HOA), implying that Runx2 inhibitors may have a potential therapeutic role in OA. Further replication studies similarly showed that Runx2 expression levels were associated with all OA and KOA, suggesting the reliability of primary findings. Unfortunately, the causal effect of Runx2 expression with HOA was not repeated in replicated studies.
OA imposes a huge economic burden on patients and society because of its associated disability and incurability [25]. Relieving pain, rather than delaying disease progression, has been the focus of clinical treatment and management of patients with OA. Researchers and clinicians have gained deeper insights into the pathogenesis of OA, and suppressing its progression through early intervention has been increasingly advocated in recent years [2]. Therefore, the development of new drug targets is urgently required. The rapid growth of GWAS and the emergence of MR analyses have fulfilled this need [17, 26]. More importantly, drug targets supported by genetic evidence have a higher potential for approval [27]. Therefore, in the present study, we used SMR analysis to examine the association between Runx2 susceptibility and OA, revealing the therapeutic potential of Runx2 inhibitors in decreasing the risk of OA.
Runx2 is an essential transcription factor in the development and maturation of the skeletal system and participates in important physiological processes, such as chondrocyte maturation and osteoblast differentiation [28]. Accumulating evidence suggests that Runx2 contributes to OA by promoting chondrocyte hypertrophy and cartilage matrix degradation [13, 29]. Notably, a recent review comprehensively illustrated the critical role of Runx2 in the pathogenesis and progression of OA and highlighted the potential of Runx2 as a therapeutic target [9]. Here, we applied SMR analysis and found that genetically predicted Runx2 expression levels were positively associated with the risk of OA; that is, Runx2 inhibitors may reduce the risk of OA. Consistent with our current findings, milder OA phenotypes were observed after the specific deletion of Runx2 in chondrocytes from surgically induced mouse OA models [7, 13, 30]. Additionally, several studies have demonstrated the benefits of Runx2 inhibition in the treatment of OA. For example, Wu et al. identified a signaling pathway in chondrocytes that exerts a protective effect on articular cartilage: kindlin-2/Stat3/Runx2 [31]. In their study, researchers demonstrated that the characteristic OA damage caused by kindlin-2 deficiency could be attenuated by deleting Runx2 in chondrocytes [31]. Yes1 associated transcriptional regulator (YAP1) was found to inhibit the transcriptional activity of Runx2, thus acting as a Runx2 inhibitor [32, 33]. Surprisingly, numerous studies have suggested that YAP1 inhibits cartilage damage, thereby preventing the subsequent deterioration of OA, suggesting that OA can be treated by activating YAP1 [34,35,36]. The vital role of microRNAs in cartilage health and OA pathogenesis has been well described [37, 38]. Interestingly, Huang et al. and Cao et al. found that treatment with exogenous microRNAs (including miR-204, miR-211, and miR-204-5p) attenuated the symptoms and delayed the progression of OA by suppressing Runx2 expression in the cartilage [14, 39]. In summary, Runx2 inhibitors exhibit great promise for the treatment of OA, and our findings provide additional evidence from a genetic perspective. Although no clinical drugs targeting Runx2 have yet been developed for the treatment of OA, we look forward to future developments.
The primary strength of our study is that we used an MR design that diminished the bias resulting from confounders and reverse causality in traditional observational studies. The evidence for causal associations was further strengthened by the success of the HEIDI test and replication study. However, this study has several limitations that must be acknowledged. First, although the GWAS summary data for OA involved in the discover study are the largest available, the participants were not exclusively of European ethnicity. This may have led to bias. Surprisingly, fewer than 3% of the participants were of non-European ancestry; thus, we believe that our results remain robust. Second, the data were mainly derived from European populations; therefore, caution should be exercised when applying our findings to other ethnicities. Finally, as no drugs targeting Runx2 have been developed, we could not establish a positive control to validate the effectiveness of the selected IV.
Conclusion
Expression levels of Runx2 were positively associated with the risk of all three OA phenotypes, implying that Runx2 inhibitors have great potential for the treatment of OA and providing new evidence from a genetic perspective. We look forward to the early development of clinical drugs targeting Runx2 to improve the well-being of patients with OA. Further MR analyses and clinical trials are required to validate these findings.
Data Availability
The GWAS summary data for OA were obtained from the Genetics of Osteoarthritis Consortium (Discovery study: https://msk.hugeamp.org/downloads.html) and UK Biobank and arcOGEN Consortium (Replication study: https://www.ebi.ac.uk/gwas/publications/30664745), respectively. Blood eQTLs data were downloaded from the eQTLGen Consortium (https://www.eqtlgen.org/cis-eqtls.html).
References
Hunter DJ, Bierma-Zeinstra S. Osteoarthritis. Lancet. 2019;393:1745–59.
Yao Q, Wu X, Tao C, et al. Osteoarthritis: pathogenic signaling pathways and therapeutic targets. Signal Transduct Target Ther. 2023;8:56.
Quicke JG, Conaghan PG, Corp N, Peat G. Osteoarthritis year in review 2021: epidemiology & therapy. Osteoarthritis Cartilage. 2022;30:196–206.
Global, regional, and national burden of osteoarthritis, 1990–2020 and projections to 2050: a systematic analysis for the Global Burden of Disease Study 2021. Lancet Rheumatol. 2023;5:e508–e522.
Fu W, Vasylyev D, Bi Y, et al. Nav1.7 as a chondrocyte regulator and therapeutic target for osteoarthritis. Nature. 2024;625:557–565.
Vargas Negrín F, Medina Abellán MD, Hermosa Hernán JC, de Felipe MR. Treatment of patients with osteoarthritis. Aten Primaria. 2014;46(Suppl 1):39–61.
Nagata K, Hojo H, Chang SH, et al. Runx2 and Runx3 differentially regulate articular chondrocytes during surgically induced osteoarthritis development. Nat Commun. 2022;13:6187.
Kim W-J, Shin H-L, Kim B-S, Kim H-J, Ryoo H-M. RUNX2-modifying enzymes: therapeutic targets for bone diseases. Exp Mol Med. 2020;52:1178–84.
Chen D, Kim DJ, Shen J, Zou Z, O’Keefe RJ. Runx2 plays a central role in osteoarthritis development. J Orthop Translat. 2020;23:132–9.
Lin AC, Seeto BL, Bartoszko JM, et al. Modulating hedgehog signaling can attenuate the severity of osteoarthritis. Nat Med. 2009;15:1421–5.
Catheline SE, Hoak D, Chang M, et al. Chondrocyte-specific RUNX2 overexpression accelerates post-traumatic osteoarthritis progression in adult mice. J Bone Miner Res. 2019;34:1676–89.
Chen J, Chen F, Wu X, et al. DLX5 promotes Col10a1 expression and chondrocyte hypertrophy and is involved in osteoarthritis progression. Genes Dis. 2023;10:2097–108.
Liao L, Zhang S, Gu J, et al. Deletion of Runx2 in articular chondrocytes decelerates the progression of DMM-induced osteoarthritis in adult mice. Sci Rep. 2017;7:2371.
Huang J, Zhao L, Fan Y, et al. The microRNAs miR-204 and miR-211 maintain joint homeostasis and protect against osteoarthritis progression. Nat Commun. 2019;10:2876.
Xie N, Xie J, Wang Z, et al. The role of calcium, 25-hydroxyvitamin D, and parathyroid hormone in irritable bowel syndrome: a bidirectional two-sample Mendelian randomization study. Nutrients. 2022;14:5109.
Zhu Z, Zhang F, Hu H, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481–7.
Reay WR, Cairns MJ. Advancing the use of genome-wide association studies for drug repurposing. Nat Rev Genet. 2021;22:658–71.
Ning Z, Huang Y, Lu H, et al. Novel drug targets for atrial fibrillation identified through Mendelian randomization analysis of the blood proteome. Cardiovasc Drugs Ther. 2023. https://doi.org/10.1007/s10557-023-07467-8.
Li Y, Sundquist K, Zhang N, Wang X, Sundquist J, Memon AA. Mitochondrial related genome-wide Mendelian randomization identifies putatively causal genes for multiple cancer types. EBioMedicine. 2023;88:104432.
Yang H, Liu D, Zhao C, et al. Mendelian randomization integrating GWAS and eQTL data revealed genes pleiotropically associated with major depressive disorder. Transl Psychiatry. 2021;11:225.
Tachmazidou I, Hatzikotoulas K, Southam L, et al. Identification of new therapeutic targets for osteoarthritis through genome-wide analyses of UK Biobank data. Nat Genet. 2019;51:230–6.
Võsa U, Claringbould A, Westra H-J, et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet. 2021;53:1300–10.
Boer CG, Hatzikotoulas K, Southam L, et al. Deciphering osteoarthritis genetics across 826,690 individuals from 9 populations. Cell. 2021;184(18):4784–18.e17.
Chen J, Ruan X, Sun Y, et al. Multi-omic insight into the molecular networks of mitochondrial dysfunction in the pathogenesis of inflammatory bowel disease. EBioMedicine. 2024;99:104934.
Liu Q, Niu J, Li H, et al. Knee symptomatic osteoarthritis, walking disability, NSAIDs use and all-cause mortality: population-based wuchuan osteoarthritis study. Sci Rep. 2017;7:3309.
Liu J, Cheng Y, Li M, Zhang Z, Li T, Luo X-J. Genome-wide Mendelian randomization identifies actionable novel drug targets for psychiatric disorders. Neuropsychopharmacology. 2023;48:270–80.
Nelson MR, Tipney H, Painter JL, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47:856–60.
Komori T. Whole aspect of Runx2 functions In skeletal development. Int J Mol Sci. 2022;21;23(10):5776.
Li F, Lu Y, Ding M, et al. Runx2 contributes to murine Col10a1 gene regulation through direct interaction with its cis-enhancer. J Bone Miner Res. 2011;26:2899–910.
Kamekura S, Kawasaki Y, Hoshi K, et al. Contribution of runt-related transcription factor 2 to the pathogenesis of osteoarthritis in mice after induction of knee joint instability. Arthritis Rheum. 2006;54:2462–70.
Wu X, Lai Y, Chen S, et al. Kindlin-2 preserves integrity of the articular cartilage to protect against osteoarthritis. Nat Aging. 2022;2:332–47.
Zaidi SK, Sullivan AJ, Medina R, et al. Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J. 2004;23:790–9.
Abou-Jaoude A, Courtes M, Badique L, et al. ShcA promotes chondrocyte hypertrophic commitment and osteoarthritis in mice through RunX2 nuclear translocation and YAP1 inactivation. Osteoarthritis Cartilage. 2022;30:1365–75.
Luo H, Yao L, Zhang Y, Li R. Liquid chromatography-mass spectrometry-based quantitative proteomics analysis reveals chondroprotective effects of astragaloside IV in interleukin-1β-induced SW1353 chondrocyte-like cells. Biomed Pharmacother. 2017;91:796–802.
Fu L, Hu Y, Song M, et al. Up-regulation of FOXD1 by YAP alleviates senescence and osteoarthritis. PLoS Biol. 2019;17:e3000201.
Deng Y, Lu J, Li W, et al. Reciprocal inhibition of YAP/TAZ and NF-κB regulates osteoarthritic cartilage degradation. Nat Commun. 2018;9:4564.
Asahara H. Current status and strategy of microRNA research for cartilage development and osteoarthritis pathogenesis. J Bone Metab. 2016;23:121–7.
Swingler TE, Wheeler G, Carmont V, et al. The expression and function of microRNAs in chondrogenesis and osteoarthritis. Arthritis Rheum. 2012;64:1909–19.
Cao J, Han X, Qi X, Jin X, Li X. miR-204-5p inhibits the occurrence and development of osteoarthritis by targeting Runx2. Int J Mol Med. 2018;42:2560–8.
Authorship
All authors involved in this study meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship, take responsibility for their respective contributions, and approve the final manuscript to be published.
Funding
This study was funded by the National Natural Science Foundation of China (No. 82072432) (to Peng Xu). The journal’s Rapid Service Fee was funded by the authors.
Author information
Authors and Affiliations
Contributions
Peng Xu and Jiale Xie contributed to the study conception and design. Jiale Xie and Xin Xu performed the primary analysis and wrote the first draft of the manuscript. Software preparation and data collection were performed by Mingyi Yang, Hui Yu, and Jinrong Hao. The first draft of the manuscript was edited by Xin Xu and Dinglong Yang. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
Jiale Xie, Xin Xu, Mingyi Yang, Hui Yu, Jinrong Hao, Dinglong Yang, and Peng Xu announce that there is no conflict of interests among them.
Ethical Approval
Data used in our study were publicly available, each original study was approved by the respective institutional ethics review board, and informed consent was obtained from the participants. Therefore, further ethical approval was not required.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Xie, J., Xu, X., Yang, M. et al. New Insights on the Therapeutic Potential of Runt-Related Transcription Factor 2 for Osteoarthritis: Evidence from Mendelian Randomization. Rheumatol Ther (2024). https://doi.org/10.1007/s40744-024-00682-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s40744-024-00682-1