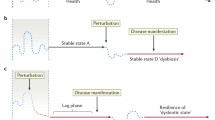
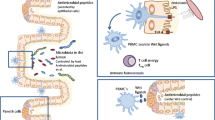
Abstract
Purpose of Review
Although extensive research has been conducted on microbial resilience, numerous unanswered questions persist. In this study, we highlight impactful research that elucidates the diverse mechanisms underlying the resilience of the gut microbiota against pathogen colonization and its implications on gut health and disease.
Recent Findings
The increasing importance of gut microbiota resistance in the context of pathogenic infections has been extensively reported. The establishment of a homeostatic microbiome-host interaction, facilitated by intricate mechanisms originating from both the microbiota and the host, plays a crucial role in fostering resilience. However, pathogens have evolved several evasion strategies that can disrupt healthy microbiota composition, trigger environmental alterations, and induce inflammation, thereby potentially exacerbating inflammatory diseases in the gut.
Summary
In this review, we aim to highlight the significance of different resilience mechanisms during intestinal infections and their potential for modulation to develop new interventions that can effectively ameliorate Inflammatory Bowel Disease (IBD).


Similar content being viewed by others
Data Availability
N/A.
Abbreviations
- IBD :
-
Inflammatory bowel disease
- SCFA :
-
Short-chain fatty acids
- MUC2 :
-
Mucin 2
- GF :
-
Germ-free
- MACs :
-
Microbiota-accessible carbohydrates
- IECs :
-
intestinal epithelial cells
- RegIIIγ :
-
Regenerating islet-derived protein IIIγ
- AMPs :
-
Antimicrobial proteins
- VRE :
-
Vancomycin-resistant enterococcus
- RIPK2 :
-
Receptor-interacting serine–threonine-protein kinase 2
- TLR :
-
Toll-like receptor
- NOD :
-
NOD-like receptor
- RLR :
-
RIG-I like receptor
- PRR :
-
Pattern-recognition receptors
- SFB :
-
Segmented filamentous bacteria
- IL :
-
Interleukin
- Th :
-
T helper
- CX3CR1 :
-
C-X3-C Motif Chemokine Receptor 1
- ILCs :
-
Innate lymphoid cells
- DSS :
-
Dextran sulfate sodium
- WT :
-
Wild-type
- SIgA :
-
Secretory immunoglobulin A
- T3SS :
-
The toxin type III secretion system
- NOX1 :
-
NADPH oxidase
- EPEC:
-
Enteropathogenic E. coli
- HNE :
-
Host nutrient extraction
- AIEC :
-
Adherent-invasive E. coli
- Kp :
-
Klebsiella pneumoniae
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Hou K, Wu ZX, Chen XY, Wang JQ, Zhang D, Xiao C, et al. Microbiota in health and diseases. Signal Transduct Target Ther. 2022;7(1):135. https://doi.org/10.1038/s41392-022-00974-4.
Berg G, Rybakova D, Fischer D, Cernava T, Verges MC, Charles T, et al. Microbiome definition re-visited: old concepts and new challenges. Microbiome. 2020;8(1):103. https://doi.org/10.1186/s40168-020-00875-0.
Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, et al. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms. 2019;7(1). https://doi.org/10.3390/microorganisms7010014.
Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J. 2017;474(11):1823–36. https://doi.org/10.1042/BCJ20160510.
Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488(7413):621–6. https://doi.org/10.1038/nature11400.
Magzal F, Shochat T, Haimov I, Tamir S, Asraf K, Tuchner-Arieli M, et al. Increased physical activity improves gut microbiota composition and reduces short-chain fatty acid concentrations in older adults with insomnia. Sci Rep. 2022;12(1):2265. https://doi.org/10.1038/s41598-022-05099-w.
Dogra SK, Dore J, Damak S. Gut Microbiota Resilience: Definition, Link to Health and Strategies for Intervention. Front Microbiol. 2020;11:572921. https://doi.org/10.3389/fmicb.2020.572921.
Wilkins LJ, Monga M, Miller AW. Defining Dysbiosis for a Cluster of Chronic Diseases. Sci Rep. 2019;9(1):12918. https://doi.org/10.1038/s41598-019-49452-y.
Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4554–61. https://doi.org/10.1073/pnas.1000087107.
Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol. 2017;15(10):630–8. https://doi.org/10.1038/nrmicro.2017.58.
Hryckowian AJ, Van Treuren W, Smits SA, Davis NM, Gardner JO, Bouley DM, et al. Microbiota-accessible carbohydrates suppress Clostridium difficile infection in a murine model. Nat Microbiol. 2018;3(6):662–9. https://doi.org/10.1038/s41564-018-0150-6.
Nguyen J, Pepin DM, Tropini C. Cause or effect? The spatial organization of pathogens and the gut microbiota in disease. Microbes Infect. 2021;23(6–7):104815. https://doi.org/10.1016/j.micinf.2021.104815.
Tropini C, Earle KA, Huang KC, Sonnenburg JL. The Gut Microbiome: Connecting Spatial Organization to Function. Cell Host Microbe. 2017;21(4):433–42. https://doi.org/10.1016/j.chom.2017.03.010.
Gu S, Chen D, Zhang JN, Lv X, Wang K, Duan LP, et al. Bacterial community mapping of the mouse gastrointestinal tract. PLoS ONE. 2013;8(10):e74957. https://doi.org/10.1371/journal.pone.0074957.
Saffarian A, Mulet C, Regnault B, Amiot A, Tran-Van-Nhieu J, Ravel J, et al. Crypt- and Mucosa-Associated Core Microbiotas in Humans and Their Alteration in Colon Cancer Patients. mBio. 2019;10(4). https://doi.org/10.1128/mBio.01315-19.
Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I, et al. Gut microbiota functions: metabolism of nutrients and other food components. Eur J Nutr. 2018;57(1):1–24. https://doi.org/10.1007/s00394-017-1445-8.
Momose Y, Hirayama K, Itoh K. Competition for proline between indigenous Escherichia coli and E. coli O157:H7 in gnotobiotic mice associated with infant intestinal microbiota and its contribution to the colonization resistance against E. coli O157:H7. Antonie Van Leeuwenhoek. 2008;94(2):165–71. https://doi.org/10.1007/s10482-008-9222-6.
Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, et al. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun. 2008;76(3):1143–52. https://doi.org/10.1128/IAI.01386-07.
Leatham MP, Banerjee S, Autieri SM, Mercado-Lubo R, Conway T, Cohen PS. Precolonized human commensal Escherichia coli strains serve as a barrier to E. coli O157:H7 growth in the streptomycin-treated mouse intestine. Infect Immun. 2009;77(7):2876–86. https://doi.org/10.1128/IAI.00059-09.
Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336(6086):1325–9. https://doi.org/10.1126/science.1222195.
Scott KP, Gratz SW, Sheridan PO, Flint HJ, Duncan SH. The influence of diet on the gut microbiota. Pharmacol Res. 2013;69(1):52–60. https://doi.org/10.1016/j.phrs.2012.10.020.
Lamichhane S, Sen P, Dickens AM, Oresic M, Bertram HC. Gut metabolome meets microbiome: A methodological perspective to understand the relationship between host and microbe. Methods. 2018;149:3–12. https://doi.org/10.1016/j.ymeth.2018.04.029.
Ruan W, Engevik MA, Spinler JK, Versalovic J. Healthy Human Gastrointestinal Microbiome: Composition and Function After a Decade of Exploration. Dig Dis Sci. 2020;65(3):695–705. https://doi.org/10.1007/s10620-020-06118-4.
Alakomi HL, Skytta E, Saarela M, Mattila-Sandholm T, Latva-Kala K, Helander IM. Lactic acid permeabilizes gram-negative bacteria by disrupting the outer membrane. Appl Environ Microbiol. 2000;66(5):2001–5. https://doi.org/10.1128/AEM.66.5.2001-2005.2000.
Knaus UG, Hertzberger R, Pircalabioru GG, Yousefi SP, Branco Dos Santos F. Pathogen control at the intestinal mucosa - H(2)O(2) to the rescue. Gut Microbes. 2017;8(1):67–74. https://doi.org/10.1080/19490976.2017.1279378.
Atassi F, Servin AL. Individual and co-operative roles of lactic acid and hydrogen peroxide in the killing activity of enteric strain Lactobacillus johnsonii NCC933 and vaginal strain Lactobacillus gasseri KS120.1 against enteric, uropathogenic and vaginosis-associated pathogens. FEMS Microbiol Lett. 2010;304(1):29–38. https://doi.org/10.1111/j.1574-6968.2009.01887.x.
Okuda K, Zendo T, Sugimoto S, Iwase T, Tajima A, Yamada S, et al. Effects of bacteriocins on methicillin-resistant Staphylococcus aureus biofilm. Antimicrob Agents Chemother. 2013;57(11):5572–9. https://doi.org/10.1128/AAC.00888-13.
Marteyn B, Scorza FB, Sansonetti PJ, Tang C. Breathing life into pathogens: the influence of oxygen on bacterial virulence and host responses in the gastrointestinal tract. Cell Microbiol. 2011;13(2):171–6. https://doi.org/10.1111/j.1462-5822.2010.01549.x.
Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. 2015;17(5):662–71. https://doi.org/10.1016/j.chom.2015.03.005.
Rivera-Chavez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, et al. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe. 2016;19(4):443–54. https://doi.org/10.1016/j.chom.2016.03.004.
Litvak Y, Mon KKZ, Nguyen H, Chanthavixay G, Liou M, Velazquez EM, et al. Commensal Enterobacteriaceae Protect against Salmonella Colonization through Oxygen Competition. Cell Host Microbez. 2019;25(1):128-39 e5. https://doi.org/10.1016/j.chom.2018.12.003.
Herp S, Brugiroux S, Garzetti D, Ring D, Jochum LM, Beutler M, et al. Mucispirillum schaedleri Antagonizes Salmonella Virulence to Protect Mice against Colitis. Cell Host Microbe. 2019;25(5):681–94 e8. https://doi.org/10.1016/j.chom.2019.03.004.
Hansson GC, Johansson ME. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Gut Microbes. 2010;1(1):51–4. https://doi.org/10.4161/gmic.1.1.10470.
Van der Sluis M, De Koning BA, De Bruijn AC, Velcich A, Meijerink JP, Van Goudoever JB, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131(1):117–29. https://doi.org/10.1053/j.gastro.2006.04.020.
Velcich A, Yang W, Heyer J, Fragale A, Nicholas C, Viani S, et al. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295(5560):1726–9. https://doi.org/10.1126/science.1069094.
Bergstrom KS, Kissoon-Singh V, Gibson DL, Ma C, Montero M, Sham HP, et al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 2010;6(5):e1000902. https://doi.org/10.1371/journal.ppat.1000902.
Zarepour M, Bhullar K, Montero M, Ma C, Huang T, Velcich A, et al. The mucin Muc2 limits pathogen burdens and epithelial barrier dysfunction during Salmonella enterica serovar Typhimurium colitis. Infect Immun. 2013;81(10):3672–83. https://doi.org/10.1128/IAI.00854-13.
Zhang T, Sasabe J, Hullahalli K, Sit B, Waldor MK. Increased Listeria monocytogenes Dissemination and Altered Population Dynamics in Muc2-Deficient Mice. Infect Immun. 2021;89(4). https://doi.org/10.1128/IAI.00667-20. This work shows that Muc2 mucin plays a key role in controlling L. monocytogenes colonization, dissemination, and population dynamics.
Johansson ME, Jakobsson HE, Holmen-Larsson J, Schutte A, Ermund A, Rodriguez-Pineiro AM, et al. Normalization of Host Intestinal Mucus Layers Requires Long-Term Microbial Colonization. Cell Host Microbe. 2015;18(5):582–92. https://doi.org/10.1016/j.chom.2015.10.007.
Neumann M, Steimle A, Grant ET, Wolter M, Parrish A, Willieme S, et al. Deprivation of dietary fiber in specific-pathogen-free mice promotes susceptibility to the intestinal mucosal pathogen Citrobacter rodentium. Gut Microbes. 2021;13(1):1966263. https://doi.org/10.1080/19490976.2021.1966263. This study indicates that absence of fibers in Western-style diets impairs the mucus layer thickness and leads to disruption of epithelial barrier through changes in microbiome composition, facilitating the infection of enteropathogens.
Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell. 2016;167(5):1339-53 e21. https://doi.org/10.1016/j.cell.2016.10.043.
Schroeder BO, Birchenough GMH, Stahlman M, Arike L, Johansson MEV, Hansson GC, et al. Bifidobacteria or Fiber Protects against Diet-Induced Microbiota-Mediated Colonic Mucus Deterioration. Cell Host Microbe. 2018;23(1):27-40 e7. https://doi.org/10.1016/j.chom.2017.11.004.
Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14(3):141–53. https://doi.org/10.1038/nri3608.
Kim YS, Ho SB. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr Gastroenterol Rep. 2010;12(5):319–30. https://doi.org/10.1007/s11894-010-0131-2.
Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9(5):356–68. https://doi.org/10.1038/nrmicro2546.
Bosch TCG, Zasloff M. Antimicrobial Peptides-or How Our Ancestors Learned to Control the Microbiome. mBio. 2021;12(5):e0184721. https://doi.org/10.1128/mBio.01847-21.
Abt MC, Buffie CG, Susac B, Becattini S, Carter RA, Leiner I, et al. TLR-7 activation enhances IL-22-mediated colonization resistance against vancomycin-resistant enterococcus. Sci Transl Med. 2016;8(327):327ra25. https://doi.org/10.1126/scitranslmed.aad6663.
Waldschmitt N, Kitamoto S, Secher T, Zacharioudaki V, Boulard O, Floquet E, et al. The regenerating family member 3 beta instigates IL-17A-mediated neutrophil recruitment downstream of NOD1/2 signalling for controlling colonisation resistance independently of microbiota community structure. Gut. 2019;68(7):1190–9. https://doi.org/10.1136/gutjnl-2018-316757.
Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307(5710):731–4. https://doi.org/10.1126/science.1104911.
Abreu MT. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol. 2010;10(2):131–44. https://doi.org/10.1038/nri2707.
Elinav E, Henao-Mejia J, Flavell RA. Integrative inflammasome activity in the regulation of intestinal mucosal immune responses. Mucosal Immunol. 2013;6(1):4–13. https://doi.org/10.1038/mi.2012.115.
Broquet AH, Hirata Y, McAllister CS, Kagnoff MF. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J Immunol. 2011;186(3):1618–26. https://doi.org/10.4049/jimmunol.1002862.
Sabat R, Ouyang W, Wolk K. Therapeutic opportunities of the IL-22-IL-22R1 system. Nat Rev Drug Discov. 2014;13(1):21–38. https://doi.org/10.1038/nrd4176.
Keir M, Yi Y, Lu T, Ghilardi N. The role of IL-22 in intestinal health and disease. J Exp Med. 2020;217(3):e20192195. https://doi.org/10.1084/jem.20192195.
Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–98. https://doi.org/10.1016/j.cell.2009.09.033.
Nagao-Kitamoto H, Leslie JL, Kitamoto S, Jin C, Thomsson KA, Gillilland MG 3rd, et al. Interleukin-22-mediated host glycosylation prevents Clostridioides difficile infection by modulating the metabolic activity of the gut microbiota. Nat Med. 2020;26(4):608–17. https://doi.org/10.1038/s41591-020-0764-0. The authors demonstrated that IL-22, induced by colonization of the gut microbiota, is crucial for the prevention of C. difficile infection in human microbiota-associated (HMA) mice.
Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14(3):282–9. https://doi.org/10.1038/nm1720.
Chen J, Waddell A, Lin YD, Cantorna MT. Dysbiosis caused by vitamin D receptor deficiency confers colonization resistance to Citrobacter rodentium through modulation of innate lymphoid cells. Mucosal Immunol. 2015;8(3):618–26. https://doi.org/10.1038/mi.2014.94.
Neil JA, Matsuzawa-Ishimoto Y, Kernbauer-Holzl E, Schuster SL, Sota S, Venzon M, et al. IFN-I and IL-22 mediate protective effects of intestinal viral infection. Nat Microbiol. 2019;4(10):1737–49. https://doi.org/10.1038/s41564-019-0470-1.
Behnsen J, Jellbauer S, Wong CP, Edwards RA, George MD, Ouyang W, et al. The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity. 2014;40(2):262–73. https://doi.org/10.1016/j.immuni.2014.01.003.
Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332(6032):974–7. https://doi.org/10.1126/science.1206095.
Franchi L, Kamada N, Nakamura Y, Burberry A, Kuffa P, Suzuki S, et al. NLRC4-driven production of IL-1beta discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat Immunol. 2012;13(5):449–56. https://doi.org/10.1038/ni.2263.
Wu WH, Kim M, Chang LC, Assie A, Saldana-Morales FB, Zegarra-Ruiz DF, et al. Interleukin-1beta secretion induced by mucosa-associated gut commensal bacteria promotes intestinal barrier repair. Gut Microbes. 2022;14(1):2014772. https://doi.org/10.1080/19490976.2021.2014772. This study demonstrated that a commensal E. coli isolate could induce IL-1b production by mononuclear phagocytes and IL-22 by ILC3 cells, improving the epithelial barrier repair after C. rodentium infection.
Kidess E, Kleerebezem M, Brugman S. Colonizing Microbes, IL-10 and IL-22: Keeping the Peace at the Mucosal Surface. Front Microbiol. 2021;12:729053. https://doi.org/10.3389/fmicb.2021.729053.
Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482(7385):395–9. https://doi.org/10.1038/nature10772.
Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107(27):12204–9. https://doi.org/10.1073/pnas.0909122107.
O’Mahony C, Scully P, O’Mahony D, Murphy S, O’Brien F, Lyons A, et al. Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-kappaB activation. PLoS Pathog. 2008;4(8):e1000112. https://doi.org/10.1371/journal.ppat.1000112.
Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34(5):794–806. https://doi.org/10.1016/j.immuni.2011.03.021.
Maharshak N, Packey CD, Ellermann M, Manick S, Siddle JP, Huh EY, et al. Altered enteric microbiota ecology in interleukin 10-deficient mice during development and progression of intestinal inflammation. Gut Microbes. 2013;4(4):316–24. https://doi.org/10.4161/gmic.25486.
Fadlallah J, El Kafsi H, Sterlin D, Juste C, Parizot C, Dorgham K, et al. Microbial ecology perturbation in human IgA deficiency. Sci Transl Med. 2018;10(439). https://doi.org/10.1126/scitranslmed.aan1217.
Nakajima A, Vogelzang A, Maruya M, Miyajima M, Murata M, Son A, et al. IgA regulates the composition and metabolic function of gut microbiota by promoting symbiosis between bacteria. J Exp Med. 2018;215(8):2019–34. https://doi.org/10.1084/jem.20180427.
Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou WC, et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science. 2018;360(6390):795–800. https://doi.org/10.1126/science.aaq0926.
Ost KS, O’Meara TR, Stephens WZ, Chiaro T, Zhou H, Penman J, et al. Adaptive immunity induces mutualism between commensal eukaryotes. Nature. 2021;596(7870):114–8. https://doi.org/10.1038/s41586-021-03722-w.
Ducarmon QR, Zwittink RD, Hornung BVH, van Schaik W, Young VB, Kuijper EJ. Gut Microbiota and Colonization Resistance against Bacterial Enteric Infection. Microbiol Mol Biol Rev. 2019;83(3). https://doi.org/10.1128/MMBR.00007-19.
Khan I, Bai Y, Zha L, Ullah N, Ullah H, Shah SRH, et al. Mechanism of the Gut Microbiota Colonization Resistance and Enteric Pathogen Infection. Front Cell Infect Microbiol. 2021;11:716299. https://doi.org/10.3389/fcimb.2021.716299.
Caballero-Flores G, Pickard JM, Nunez G. Microbiota-mediated colonization resistance: mechanisms and regulation. Nat Rev Microbiol. 2023;21(6):347–60. https://doi.org/10.1038/s41579-022-00833-.
Lopez CA, Miller BM, Rivera-Chavez F, Velazquez EM, Byndloss MX, Chavez-Arroyo A, et al. Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science. 2016;353(6305):1249–53. https://doi.org/10.1126/science.aag3042.
Miller BM, Liou MJ, Zhang LF, Nguyen H, Litvak Y, Schorr EM, et al. Anaerobic Respiration of NOX1-Derived Hydrogen Peroxide Licenses Bacterial Growth at the Colonic Surface. Cell Host Microbe. 2020;28(6):789-97 e5. https://doi.org/10.1016/j.chom.2020.10.009. The authors describes one of the several evasion strategies of C. rodentium in which host NOX1-derived H2O2 fuels its anaerobic growth during early infection, while the pathogen relies on aerobic respiration for its expansion in the inflamed gut.
Pal RR, Baidya AK, Mamou G, Bhattacharya S, Socol Y, Kobi S, et al. Pathogenic E coli Extracts Nutrients from Infected Host Cells Utilizing Injectisome Components. Cell. 2019;177(3):683-96 e18. https://doi.org/10.1016/j.cell.2019.02.022.
Jimenez AG, Ellermann M, Abbott W, Sperandio V. Diet-derived galacturonic acid regulates virulence and intestinal colonization in enterohaemorrhagic Escherichia coli and Citrobacter rodentium. Nat Microbiol. 2020;5(2):368–78. https://doi.org/10.1038/s41564-019-0641-0.
Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2(3):204. https://doi.org/10.1016/j.chom.2007.08.002.
Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5(10):2177–89. https://doi.org/10.1371/journal.pbio.0050244.
Lopez CA, Winter SE, Rivera-Chavez F, Xavier MN, Poon V, Nuccio SP, et al. Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. mBio. 2012;3(3). https://doi.org/10.1128/mBio.00143-12.
Faber F, Tran L, Byndloss MX, Lopez CA, Velazquez EM, Kerrinnes T, et al. Host-mediated sugar oxidation promotes post-antibiotic pathogen expansion. Nature. 2016;534(7609):697–9. https://doi.org/10.1038/nature18597.
Taroncher-Oldenburg G, Jones S, Blaser M, Bonneau R, Christey P, Clemente JC, et al. Translating microbiome futures. Nat Biotechnol. 2018;36(11):1037–42. https://doi.org/10.1038/nbt.4287.
Neurath MF. Host-microbiota interactions in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17(2):76–7. https://doi.org/10.1038/s41575-019-0248-1.
Caruso R, Lo BC, Nunez G. Host-microbiota interactions in inflammatory bowel disease. Nat Rev Immunol. 2020;20(7):411–26. https://doi.org/10.1038/s41577-019-0268-7.
Schaubeck M, Clavel T, Calasan J, Lagkouvardos I, Haange SB, Jehmlich N, et al. Dysbiotic gut microbiota causes transmissible Crohn’s disease-like ileitis independent of failure in antimicrobial defence. Gut. 2016;65(2):225–37. https://doi.org/10.1136/gutjnl-2015-309333.
Elhenawy W, Hordienko S, Gould S, Oberc AM, Tsai CN, Hubbard TP, et al. High-throughput fitness screening and transcriptomics identify a role for a type IV secretion system in the pathogenesis of Crohn’s disease-associated Escherichia coli. Nat Commun. 2021;12(1):2032. https://doi.org/10.1038/s41467-021-22306-w. This study reveals the role of type IV secretion system in biofilm formation by adherent-invasive E. coli isolated from Crohn’s disease patients, which was essential in promoting its persistence in the gut.
Seishima J, Iida N, Kitamura K, Yutani M, Wang Z, Seki A, et al. Gut-derived Enterococcus faecium from ulcerative colitis patients promotes colitis in a genetically susceptible mouse host. Genome Biol. 2019;20(1):252. https://doi.org/10.1186/s13059-019-1879-9.
Zamani S, HesamShariati S, Zali MR, AsadzadehAghdaei H, SarabiAsiabar A, Bokaie S, et al. Detection of enterotoxigenic Bacteroides fragilis in patients with ulcerative colitis. Gut Pathog. 2017;9:53. https://doi.org/10.1186/s13099-017-0202-0.
Kitamoto S, Nagao-Kitamoto H, Jiao Y, Gillilland MG 3rd, Hayashi A, Imai J, et al. The Intermucosal Connection between the Mouth and Gut in Commensal Pathobiont-Driven Colitis. Cell. 2020;182(2):447-62 e14. https://doi.org/10.1016/j.cell.2020.05.048. This study reported that periodontitis leads to an expansion of oral pathobionts enabling its translocation into gut, where they activate inflammasomes of colonic mononuclear phagocytes and triggering intestinal inflammation.
Capurso L. Thirty Years of Lactobacillus rhamnosus GG: A Review. J Clin Gastroenterol. 2019;53(Suppl 1):S1–41. https://doi.org/10.1097/MCG.0000000000001170.
Karczewski J, Troost FJ, Konings I, Dekker J, Kleerebezem M, Brummer RJ, et al. Regulation of human epithelial tight junction proteins by Lactobacillus plantarum in vivo and protective effects on the epithelial barrier. Am J Physiol Gastrointest Liver Physiol. 2010;298(6):G851–9. https://doi.org/10.1152/ajpgi.00327.2009.
Federici S, Kredo-Russo S, Valdes-Mas R, Kviatcovsky D, Weinstock E, Matiuhin Y, et al. Targeted suppression of human IBD-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation. Cell. 2022;185(16):2879-98 e24. https://doi.org/10.1016/j.cell.2022.07.003. In this study the authors developed a feasible oral administration of a phage therapy targeting IBD-associated Klebsiella pneumoniae that contributes to amelioration of the disease.
Funding
MC, EMS and NOSC are supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) with grant numbers 2018/24350–4, 2020/14388–4 and 2017/05264–7, respectively.
Author information
Authors and Affiliations
Contributions
MC: Writing—original draft, figure preparation; writing—review and editing.
EMS: Writing—original draft; writing—review and editing.
NOSC: Conceptualization, lead; funding acquisition, supporting; lead; writing—original draft, lead; writing—review and editing, lead.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Topical Collection on Microbe-Microbiome Interactions
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cipelli, M., da Silva, E.M. & Câmara, N.O.S. Gut Microbiota Resilience Mechanisms Against Pathogen Infection and its Role in Inflammatory Bowel Disease. Curr Clin Micro Rpt 10, 187–197 (2023). https://doi.org/10.1007/s40588-023-00207-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40588-023-00207-4