Abstract
The increased availability of digital data from electronic health records (EHR) systems has created heightened interest in their use in pharmacoepidemiology. Sources of such data have been somewhat restrictive and not generally accessible to scientists because of institutional policies. However, these data-access restrictions appear to be diminishing. The recent thrust by the Institute of Medicine and other policy-influencing groups to establish learning health systems has argued for broader use of digital data from health systems for the growing needs of researchers who seek out best evidence in health care. Pharmacoepidemiologists are especially well poised to contribute to this new era because of their long-standing use of digital data in their research. While EHR data will increase in terms of volume, it is important for investigators to spend time understanding the data, including verifying format and quality. Unlike claims data that often follow a standard format, EHR data, particularly from disparate health systems such as contained in health information exchanges, often vary in terms of completeness, format, and quality. It is therefore helpful for investigators to work closely with individuals who are tuned into each data source being considered for research. EHR data are at the core of the exciting new thrust to analyze big healthcare data for pharmacoepidemiology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Data from electronic health records (EHRs) are ubiquitous and increasingly used for research in clinical epidemiology, particularly pharmacoepidemiology. A distinction can be made between an EHR and an EMR (or electronic medical record), although these terms are often used interchangeably. The EMR is basically a practice-based digital medical record with data that often does not transfer out of the practice very well. In contrast, an EHR contains data external to the practice’s medical record per se, such as data from external laboratories or patient portals, and can be shared beyond the practice site [1]. Recent estimates indicate that nine out of ten health systems have adopted an EHR, although only a third of these systems meet the basic criteria for the Centers for Medicare and Medicaid Services meaningful use Stage 1 requirements [2] that include electronic ordering of prescriptions [3]. However, while capturing physician order data may not be available for research, access to dispensing data often is. Pharmacy departments have been early adopters of computerization, largely because maintaining prescription dispensing records with paper prescriptions is especially onerous. Therefore, even these basic systems collect large digital repositories of prescription data useful for pharmacoepidemiology and pharmacovigilance. More advanced systems are now being tapped for participation in major efforts in the US to use electronic health records and administrative data, including shared networks, for pharmacoepidemiology, such as the Food and Drug Administration’s Mini-Sentinel network [4].
There have been several recent reviews of the application of the methods of pharmacoepidemiology using data from EHRs [5–7]. Often missing from these reports is the perspective of the stewards of these digital repositories from institutional EHRs and health information exchanges that incorporate data from multiple institutions [8]. Understanding this perspective is important in study design, for assessing data limitations, and to ensure accurate interpretation of data from electronic data sources; that is, translating data into useful information. As such, the purpose of this report is to describe important considerations in the use of EHR data for research from the perspective of the stewards of one of the oldest longitudinal electronic medical record systems: namely, the Regenstrief Institute.
Digital Data: From a Small Clinic to a Health Information Exchange
Regenstrief Institute began working with electronic healthcare data in 1972 with the creation of the Regenstrief Medical Record System [9]. The system was first established by Dr Clement McDonald in a medicine subspecialty clinic serving patients with diabetes at Wishard Health Services in Indianapolis, Indiana, USA. Other clinics were progressively added along with Wishard’s hospital and emergency department. The structure of the system was modular with registration and scheduling, appointments, laboratory, pharmacy, and procedure modules that fed a central archival database that could be queried using a cryptic retrieval language called CARE. This archival database was the source of data for early projects by a number of renowned clinical epidemiologists including Drs Robert Dittus (Vanderbilt University), Bruce Psaty (Washington University), and William Tierney (Indiana University), who often wrote their own CARE programs to extract data from the database. As demand for the clinical data grew, a group of data managers emerged to satisfy researcher demand and to serve the quality improvement needs of Wishard [10].
Other healthcare institutions throughout Indianapolis established EHRs through the 1990s. It became apparent that there were opportunities to share patient data for patients who received care at multiple facilities and by doing so eliminate the costs of duplicate care and assessments such as multiple expensive radiology scans. To address this need, the Indianapolis Network for Patient Care was established in 1995 as a federation of data repositories wherein each healthcare institution throughout Indianapolis retained ownership of its data but shared them as needed to support care [11]. Over time, a growing number of other facilities throughout the state of Indiana participated and in 2004 the Indiana Network for Patient Care (INPC) was established to provide clinical data services for its members whose data were contained in the Indiana Health Information Exchange (www.ihie.org). The exchange currently houses a centralized database that contains digital data from more than 100 healthcare systems throughout Indiana. Regenstrief Institute oversees a derivative copy of this database (INPC-R) that serves the needs of quality improvement and research. This database contains medical record data for 14.7 million unique patients but data vary considerably in their density. Notably, patients in facilities more recently added to the INPC have fewer data available and even the types of data exchanged may vary. Collectively, however, there are 34.1 million registration events, 4.7 billion clinical observations, 776 million claims including prescriptions, and 136.8 million text reports.
The Regenstrief Institute Data Core
Because of the growing data needs of researchers, a data core was established at Regenstrief Institute that currently has 13 Masters and PhD-level analysts. In any given week, there are 50–60 ongoing research projects involving data extracted from the INPC-R. A key advantage of this group of analysts is that they have an understanding of how EHR data move from the clinical arena into the INPC-R from their routine work with health system physicians and staff. Another advantage is that the analysts work as a team and share solutions to problems and other experiences while serving the needs of researchers.
Data from Electronic Health Records (EHRs)
Table 1 compares various data types found in the electronic health data contained in the INPC-R and in administrative claims data. EHR data are rich but have greater variability in availability among data types. Unlike claims data, which follow a common structure, much of the data from the EHR is found in clinical notes and extracted using natural language processing (NLP) and only then may be transformed into structured or coded variables. A major gap for pharmacoepidemiologists is that symptom, side effect and adverse drug event data are predominantly found in these clinical notes. Depending on the data source, prescription data may be more complete for claims data than EHR data. For example, claims are the payment schema between healthcare providers and insurers so prescription claims are available across providers and disparate healthcare systems. However, a single health system’s EMR may include data from only that particular system. This problem can be resolved with health information exchanges that incorporate data from health systems and insurer claims. Alternatively, a health system’s EMR may contain information about drug product samples, over-the-counter (OTC) drugs, and dietary supplements used by patients that would not be found in claims data.
Each new project using data from EHR systems constitutes a new adventure. The reason for this is that each project requires operational definitions for dependent variables and covariates. Digital definitions of diseases (phenotypes) may vary. For example, definitions for diabetes mellitus vary using ICD9 codes, laboratory data, prescription records, or all of the above [12]. When only administrative data are available, the investigator may be limited to using only diagnostic codes and drug dispensing data. Data from the EHR may extend the digital definition to include laboratory data such as hemoglobin A1c and blood glucose results.
Pharmacoepidemiology Using EHR Data
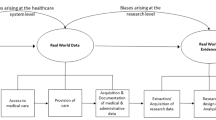
Figure 1 shows a typical pathway of issues that an investigator needs to tread in project planning. For any given research question or problem, the investigator decides the best study design and then determines what data elements from an EHR are needed and which EHR data source contains those data. Communication with the data stewards for the EHR is an important step in assessing the validity, reliability, and completeness of the data [4, 13]. If the preferred EHR data source appears inaccessible, partnering with investigators from the data source may improve data access. Part of the assessment process involves determining whether the EHR data accommodate the preferred research design and whether the data are affordable. Most data sources must pay for the data managers who extract system data for research even in nonprofit organizations. On analysis it again helps to have access to team members at the level of the data source or the data stewards to clarify what is likely to be several questions about the data, and these individuals will likely be able to assist with the interpretation of the results.
It helps considerably if the investigators have at least a minimal understanding of the data they are receiving for analysis. Data provenance (ownership and historical course of data) is available for data within the INPC-R but may be lost with the deidentification process unless the Institutional Review Board (IRB) permits the data core to retain a copy of the data set complete with identifiers and the source of data. This process is particularly important for projects using data from a multisource health information exchange. Similar data elements may appear to measure the same variable but could have a different dictionary term name or code [14]. Data mapping on the front end of the project can reduce the confusion around variable creation but sometimes the project requires verification of a variable at analysis, including provenance. Given the potential for data variability across systems, it is advisable to include the data source/health system as a stratification variable.
Each data use instance requires a specific data extraction, transformation, and loading (ETL) routine. For example, the work necessary to create an Informatics for Integrating Biology and the Bedside (I2B2) [15] instance differs significantly from that creating an Observational Medical Outcomes Partnership common data model. The time and attention of software engineers and programmers and other resources to create various databases is important. Coded or structured data from EHRs is easiest to understand and analyze. Unfortunately, much of the data contained within EHRs are text that require NLP software to extract or re-code [16]. Symptom data, which is often of interest to pharmacoepidemiologists studying adverse drug events, are often found in clinician’s notes. Similarly, unearthing functional status data such as the New York Heart Association (NYHA) classification from medical records often requires NLP. Software engineers have recently developed an open-source tool that accurately probes digital text records to find medications with great precision [17].
Attention to the project team’s membership is important [13, 18]. Unless the principal investigator is a content expert in the research interest area, access to a clinician with content expertise (internal medicine, cardiology, endocrinology) is especially helpful in understanding the types of medications prescribed to treat a particular condition, the range of dosages, and what alternative treatments (including OTC drugs and dietary supplements such as herbal medications) patients may use. Further, because study design and analysis are critical to the project’s success, an epidemiologist and/or biostatistician will make important contributions. More complex phenotypes such as drug-induced liver disease could require biomedical informatician expertise [19, 20].
Linking Data Across Healthcare Facilities
The ability to link and merge data is an important requisite of the EHR and is essential to advances in the information sciences in health care such as the conduct of observational research [21] and large simple trials [22]. Practically all future opportunities to improve health care of individuals and the overall health of the population, and reduce the costs of care are dependent on the ability to integrate data [23]. Standard formats for data elements are necessary for coding, structuring, and sharing data [23–26]. Basic structured data generally follows a standard format of date, name, and a value. For example, a prescription record generally contains the date of dispensing, the drug product name, and the product’s dosage with pointers to other coded identifiers such as the National Drug Code (NDC) that bundles product information, strength, units of strength, quantity dispensed, directions for use (often as text), and cost, among others. In the US, the open source RxNorm is a common standard format for medication data as are commercially available software from Medi-Span®; however, the World Health Organization’s Anatomical, Therapeutic and Chemical classification system and the Defined Daily Dose (ATC/DDD) Index has broad international use [27]. Similarly, laboratory and clinical data follow standard conventions by Logical Observation Identifiers Names and Codes (LOINC®) or Systematized Nomenclature of Medicine–Clinical Terms (SNOMED-CT) [28]. Data that do not follow a standard format must undergo an onerous mapping process to be useful in research. Mapping is particularly problematic when data derive from EHRs at multiple health systems: The same variable may be named differently in each system [14], which can result in missing or erroneous data.
As mentioned previously, patients whose data reside in the INPC-R may have encounters at multiple acute or primary care and referral care facilities. Each of these facilities has its own medical record assignment for each patient and sometimes the same institution may have multiple medical record numbers for the same individual. Therefore, a special probabilistic algorithm was developed by Grannis and colleagues to integrate the multiple medical record numbers for each patient into a single global patient identifier that links all data for a distinct individual within and across care sites [29, 30]. This mechanism, with few exceptions, eliminates the need for human adjudication by using an expectation maximization algorithm to find one true medical record with a sensitivity of ≥ 99%. The original algorithm used patient’s last name, transformations on the first name, middle initial, gender, and the month, day, and year of birth; however, there are ongoing revisions to the algorithm to further improve its performance.
Data Ownership
Trust building is a key factor in the use of clinical data from various healthcare providers for the purpose of research. Regenstrief Institute does not own the data contained in the INPC-R. Instead, it acts as a data steward by contract with each health system participating in the INPC. A representative from each facility participates on the INPC Management Board, which approves all research projects conducted using data from participating healthcare facilities. Gaining the trust of the public is also an important aspect of the use of medical record data for research, especially as it relates to the heightened interest in the learning health system. The goal of this system set by the Institute of Medicine (IOM) is that by 2020, 90% of decisions in health care will use timely clinical information reflecting the best evidence [31]. Clearly, accomplishing this goal will require access to routinely updated data from EHRs. Patients, providers, and health professionals must trust that digital healthcare data are used appropriately to achieve this goal [32]. The hope is that by improving access to electronic medical record data and providing care with the best evidence, health care will improve for individual patients and the costs of care will decrease. The IOM also recognizes the importance of improved privacy and confidentiality regulation aimed at reducing the burden of gaining access to important data that accelerates healthcare improvement and discovery [33]. The results of recent IOM forums and workshops have highlighted the relevance of the learning health system to observational research and trials and access to digital healthcare data will likely improve [21, 22, 32].
Preliminary data to determine the prospects of a full study are available without IRB approval; however, the IRB approves research after the preliminary phase. Multicenter studies may require IRB approval by each site or agreement among the sites to accept one center’s approval. Upon approval of the research protocol by the IRB, project data may pass from a data manager to the principal investigator or an approved member of their team such as a biostatistician. Data validity must be examined and any questions around the quality of the data addressed. Data analysis proceeds with interpretation of the results. Often after analysis begins, additional variables are requested, which in turn may require an amendment to the protocol and IRB. Upon completion of the project and publication of the results, the data may need to be returned to the data steward or destroyed. At Regenstrief Institute, data managers keep a secured file containing the data and associated documents for 7 years in case there is a future need (e.g., addressing reviewer’s comments on a manuscript involving the data or a request for data for a meta-analysis), but this time period can be shorter or longer depending on the nature of the project. At times, investigators will want to do another project from a pre-existing data set in their possession. However, it should be kept in mind that there may be many more recent and complete data available for analysis and that IRB restrictions may prohibit re-use of the pre-existing data. Therefore, a new data extraction might be advisable.
Common Misconceptions in the Use of EHR Data
A common problem new investigators sometimes have is that they confuse a database capturing the health care of people with a population database. Several countries in Europe have well defined populations with unique identifiers for individuals whose health information is captured almost completely over time. However, data from an EHR captures data primarily for disease prevention and treatment. Patient encounters with the healthcare system occur by appointment or an urgent visit for active illness. Also, encounters may occur at various settings of care that are not captured, such as out-of-system care and pharmacy-based clinics. Because measurements contained in EHRs are often from sick patients, and unless the patient returns in good health for repeat measurement, such results are not representative of the normal state of health.
It is sometimes assumed that data from EHRs are easily extracted and transformed into useful variables for research. However, there may be limitations accessing comprehensive real-time data because of their size and storage needs, the ability to process these data in real time, and interference with production uses when data extractions for research are performed in the same computing environment concurrent with clinical care. While data storage and processing speeds historically were major problems in pharmacoepidemiology and clinical research using EHRs [18], improved digital processing and storage technologies have reduced these problems and their associated costs considerably. Finally, EHR data are noisy: data are often missing, appear at irregular intervals, and may not be sufficiently structured for the research being considered. As such, the quality of analytic data from EHRs and the variables created from such data should undergo validation so that only high-quality data are analyzed and interpreted.
Future Directions
Big data has become a focus of interest in pharmacoepidemiology. Enormous databases result with the incorporation of digital data from EHRs alone but can be especially overwhelming, for example, with the addition of genomic data. Further, there is a growing interest in merging data from EHRs with claims data (e.g., Medicare) and patient-centered surveys (potentially from mobile devices) for patient-reported outcomes including health-related quality of life [34–36], which is often unavailable within an EHR. Small internal validation studies are often considered in a big data study to assess confounding variables. These smaller validation studies are less onerous and can provide valuable information to the investigation team at more manageable costs (in terms of resource utilization) than working with the entire big dataset. Investigators have been interested in capturing purchasing data or data from pharmacy loyalty card programs, which may have information on the purchases of OTC drugs or dietary supplements, and helpful products such as exercise equipment or potentially harmful substances such as cigarettes. While it may be difficult to merge data from social media sources into the EHR, these data sources have nonetheless attracted the attention of big data analysts [37].
Large comprehensive databases will require new methods for analysis and visualization. Such methods promote speed of processing and ease of comprehension of underlying patterns of data [38]. While the notion of data mining produces mixed reactions from traditional epidemiologists, biostatisticians, and other clinical scientists, a variety of new tools are evolving for drug discovery and repurposing. It is also hoped that the use of ontologies will facilitate variable construction and knowledge representation [39]. Importantly, if the mandate for implementation of ICD-10 in 2015 holds, analysts might prepare themselves for some shifts in the prevalence and incidence of some disorders as coders undergo transition to the new codes.
Finally, training a future workforce to analyze large EHR databases is critical. While the costs of training and equipping the new cadre of big data analysts will be substantial, the missed opportunity costs of not doing so will likely be even greater. One estimate by McKinsey Global Institute suggests that in the US, the availability and careful analysis of big healthcare data would have a per annum value of US$300 to US$450 billion [40]. If this estimate holds true, a small fraction of such savings could support the training of many new big data analysts in pharmacoepidemiology.
Conclusion
Data from EHRs are increasingly seen as a key aspect to the future of pharmacoepidemiology, pharmacovigilance, and other clinical research requiring observational clinical data including registries and large simple clinical trials. The growing volume of clinical data contained in EHRs must be made readily available at a reasonable cost with careful attention to privacy and confidentiality concerns of health providers and patients. There are many examples of productive use of data from EHRs that have resulted in improved care and healthcare cost reductions. Such cases are especially important to engage the greater healthcare community of providers, insurers, regulators, and governmental agencies. Barriers to accessing data from EHRs need to be quickly surmounted in order for needed healthcare improvement at the individual patient and population levels with the associated cost savings. In this respect, drug effectiveness and safety will always be an important aspect of this driving interest. New analytical methods, NLP to capture needed data from clinical notes for missing variables, and big data visualization technologies will be welcome tools in bolstering pharmacoepidemiology and pharmacovigilance using data from EHRs [41, 42, 13].
References
Garrett P, Seidman J. 2011. Retrieved August 19, 2014, 2014, from http://www.healthit.gov/buzz-blog/electronic-health-and-medical-records/emr-vs-ehr-difference/.
Jones EB, Furukawa MF. Adoption and use of electronic health records among federally qualified health centers grew substantially during 2010-12. Health Aff (Millwood). 2014;33:1154–61.
Blumenthal D, Tavenner M. The "meaningful use" regulation for electronic health records. N Engl J Med. 2010;363:501–4.
Curtis LH, Brown J, Platt R. Four health data networks illustrate the potential for a shared national multipurpose big-data network. Health Aff (Millwood). 2014;33:1178–86.
Ray WA. Improving automated database studies. Epidemiology. 2011;22:302–4.
Schneeweiss S, Avorn J. A review of uses of health care utilization databases for epidemiologic research on therapeutics. J Clin Epidemiol. 2005;58:323–37.
Yasmina A, Deneer VH, van der Zee AH, van Staa TP, de Boer A, Klungel OH. Application of routine electronic health record databases for pharmacogenetic research. J Intern Med. 2014.
Hripcsak G, Bloomrosen M, FlatelyBrennan P, et al. Health data use, stewardship, and governance: ongoing gaps and challenges: a report from AMIA's 2012 Health Policy Meeting. J Am Med Inform Assoc. 2014;21:204–11.
McDonald CJ, Overhage JM, Tierney WM, et al. The Regenstrief Medical Record System: A quarter century experience. Int J Med Inform. 1999;54:225–53.
Murray MD, Smith FE, Fox J, et al. Structure, functions, and activities of a research support informatics section. J Am Med Inform Assoc. 2003;10:389–98.
McDonald CJ, Overhage JM, Barnes M, et al. The Indiana network for patient care: a working local health information infrastructure. An example of a working infrastructure collaboration that links data from five health systems and hundreds of millions of entries. Health Aff (Millwood). 2005;24:1214–20.
Richesson RL, Rusincovitch SA, Wixted D, et al. A comparison of phenotype definitions for diabetes mellitus. J Am Med Inform Assoc. 2013;20:e319–26.
Roski J, Bo-Linn GW, Andrews TA. Creating value in health care through big data: opportunities and policy implications. Health Aff (Millwood). 2014;33:1115–22.
Psaty BM, Breckenridge AM. Mini-Sentinel and regulatory science–big data rendered fit and functional. N Engl J Med. 2014;370:2165–7.
i2b2 Collaborative. i2b2: Informatics for Integrating Biology & the Bedside. Retrieved August 20, 2014, 2014, from https://www.i2b2.org/.
Capurro D, Yetisgen M, van Eaton E, Black R, Tarczy-Hornoch P. Availability of Structured and Unstructured Clinical Data for Comparative Effectiveness Research and Quality Improvement: A Multi-Site Assessment. eGEMs (Generating Evidence & Methods to improve patient outcomes). 2014;2:1, Article 11.
Sohn S, Clark C, Halgrim SR, Murphy SP, Chute CG, Liu H. MedXN: an open source medication extraction and normalization tool for clinical text. J Am Med Inform Assoc. 2014;21:858–65.
Halamka JD. Early experiences with big data at an academic medical center. Health Aff (Millwood). 2014;33:1132–8.
Shin J, Hunt CM, Suzuki A, Papay JI, Beach KJ, Cheetham TC. Characterizing phenotypes and outcomes of drug-associated liver injury using electronic medical record data. Pharmacoepidemiol Drug Saf. 2013;22:190–8.
Overby CL, Pathak J, Gottesman O, et al. A collaborative approach to developing an electronic health record phenotyping algorithm for drug-induced liver injury. J Am Med Inform Assoc. 2013;20:e243–52. doi:10.1136/amiajnl-2013-001930.
Institute of Medicine. Observational Studies in a Learning Health System: Workshop Summary. The Learning Health System Series. Washington, DC: National Academies Press; 2013.
Institute of Medicine. Large simple trials and knowledge generation in a learning health system: Workshop Summary. The Learning Health System Series. Washington, DC: National Academies Press; 2013.
Bates DW, Saria S, Ohno-Machado L, Shah A, Escobar G. Big data in health care: using analytics to identify and manage high-risk and high-cost patients. Health Aff (Millwood). 2014;33:1123–31.
McDonald CJ, Overhage JM, Dexter P, Takesue B, Suico JG. What is done, what is needed and what is realistic to expect from medical informatics standards. Int J Med Inform. 1998;48:5–12.
Kush R, Goldman M. Fostering responsible data sharing through standards. N Engl J Med. 2014;370:2163–5.
McDonald CJ, Hammond WE. Standard formats for electronic transfer of clinical data. Ann Intern Med. 1989;110:333–5.
World Health Organization. ATC/DDD Index 2014. Retrieved August 20, 2014, 2014, from http://www.whocc.no/atc_ddd_index/.
Hammond WE, Richesson RL. Standards Development and the Future of Research Data Sources, Interoperability, and Exchange. In: Richesson RL, Andrews JE, editors. Clincial Research Informatics. London: Springer; 2012. p. 335–65.
Grannis SJ, Overhage JM, Hui S, McDonald CJ. Analysis of a probabilistic record linkage technique without human review. In: AMIA Annual Sympolium Proceedings; 2003:259-263.
Zhu VJ, Overhage MJ, Egg J, Downs SM, Grannis SJ. An empiric modification to the probabilistic record linkage algorithm using frequency-based weight scaling. J Am Med Inform Assoc. 2009;16:738–45.
Institute of Medicine. Best care at lower cost: the path to continuously learning health care in America. Washington, DC: National Academies Press; 2013.
Okun S, McGraw D, Stang P, et al. Making the Case for Continuous Learning from Routinely Collected Data. Washington, DC: Institute of Medicine; 2013.
Sugarman J, Califf RM. Ethics and regulatory complexities for pragmatic clinical trials. JAMA. 311;(23):2381-2382.
Snyder CF, Jensen RE, Segal JB, Wu AW. Patient-reported outcomes (PROs): putting the patient perspective in patient-centered outcomes research. Med Care. 2013;51 suppl 3:S73–9.
Wu AW, Kharrazi H, Boulware LE, Snyder CF. Measure once, cut twice–adding patient-reported outcome measures to the electronic health record for comparative effectiveness research. J Clin Epidemiol. 2013;66 suppl 8:S12–20.
Krist AH, Woolf SH. A vision for patient-centered health information systems. JAMA. 2011;305:300–1.
Weber GM, Mandl KD, Kohane IS. Finding the Missing Link for Big Biomedical Data. JAMA. 2014.
Schneeweiss S. Learning from big health care data. N Engl J Med. 2014;370:2161–3.
Fung KW, Bodenreider O. Knowledge Representation and Ontologies. In: Richesson RL, Andrews JE, editors. Clincial Research Informatics. London: Springer; 2012. p. 255–75.
Manyika J, Chui M, Farrell D, Van Kuiken S, Groves P, Doshi EA. Open data: Unlocking innovation and performance with liquid information. New York: McKinsey & Company; 2013.
Duke JD, Li X, Grannis SJ. Data visualization speeds review of potential adverse drug events in patients on multiple medications. J Biomed Inform. 2010;43:326–31.
Harmark L, van Grootheest AC. Pharmacovigilance: methods, recent developments and future perspectives. Eur J Clin Pharmacol. 2008;64:743–52.
Acknowledgments
The author thanks Ms. Cindy Majors for her assistance organizing references and formatting this manuscript.
Compliance with Ethics Guidelines
ᅟ
Conflict of Interest
MD Murray declares no conflicts of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Murray, M.D. Use of Data from Electronic Health Records for Pharmacoepidemiology. Curr Epidemiol Rep 1, 186–193 (2014). https://doi.org/10.1007/s40471-014-0020-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40471-014-0020-6