Abstract
Fetal growth restriction is a pathological condition occurring when the fetus does not reach the genetically determined growth potential. The etiology of fetal growth restriction is expected to be multifactorial and include fetal, maternal, and placental factors, the latter being the most frequent cause of isolated fetal growth restriction. Severe fetal growth restriction has been related to both an increased risk of perinatal morbidity and mortality, and also a greater susceptibility to developing diseases (especially cardio-metabolic and neurological disorders) later in life. In the last decade, emerging evidence has supported the hypothesis of the Developmental Origin of Health and Disease, which states that individual developmental ‘programming’ takes place via a delicate fine tuning of fetal genetic and epigenetic marks in response to a large variety of ‘stressor’ exposures during pregnancy. As the placenta is the maternal-fetal interface, it has a crucial role in fetal programming, such that any perturbation altering placental function interferes with both in-utero fetal growth and also with the adult life phenotype. Several epigenetic mechanisms have been highlighted in modulating the dynamic placental epigenome, including alterations in DNA methylation status, post-translational modification of histones, and non-coding RNAs. This review aims to provide a comprehensive and critical overview of the available literature on the epigenetic background of fetal growth restriction. A targeted research strategy was performed using PubMed, MEDLINE, Embase, and The Cochrane Library up to January 2022. A detailed and fully referenced synthesis of available literature following the Scale for the Assessment of Narrative Review Articles guidelines is provided. A variety of epigenetic marks predominantly interfering with placental development, function, and metabolism were found to be potentially associated with fetal growth restriction. Available evidence on the role of environmental exposures in shaping the placental epigenome and the fetal phenotype were also critically discussed. Because of the highly dynamic crosstalk between epigenetic mechanisms and the extra level of complexity in interpreting the final placental transcriptome, a full comprehension of these phenomenon is still lacking and advances in multi-omics approaches are urgently needed. Elucidating the role of epigenetics in the developmental origins of health and disease represents a new challenge for the coming years, with the goal of providing early interventions and prevention strategies and, hopefully, new treatment opportunities.

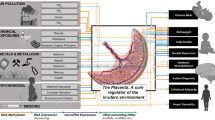
Adapted from Goyal et al. [14]
Similar content being viewed by others
References
Salafia CM, Charles AK, Maas EM. Placenta and fetal growth restriction. Clin Obstet Gynecol. 2006;49:236–56.
Gordijn SJ, Beune IM, Thilaganathan B, Papageorghiou A, Baschat AA, Baker PN, et al. Consensus definition of fetal growth restriction: a Delphi procedure. Ultrasound Obstet Gynecol. 2016;48:333–9.
ACOG practice bulletin No. 227: fetal growth restriction. Obstet Gynecol. 2021;137:e16–28.
Sun C, Groom KM, Oyston C, Chamley LW, Clark AR, James JL. The placenta in fetal growth restriction: what is going wrong? Placenta. 2020;96:10–8.
Lackman F, Capewell V, Gagnon R, Richardson B. Fetal umbilical cord oxygen values and birth to placental weight ratio in relation to size at birth. Am J Obstet Gynecol. 2001;185:674–82.
Figueras F, Gratacos E. An integrated approach to fetal growth restriction. Best Pract Res Clin Obstet Gynaecol. 2017;38:48–58.
Hemberger M. Epigenetic landscape required for placental development. Cell Mol Life Sci. 2007;64:2422–36.
Waddington CH. The epigenotype. 1942. Int J Epidemiol. 2012;41:10–3.
Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–32.
Bird A. Perceptions of epigenetics. Nature. 2007;447:396–8.
Lappalainen T, Greally JM. Associating cellular epigenetic models with human phenotypes. Nat Rev Genet. 2017;18:441–51.
Skvortsova K, Iovino N, Bogdanović O. Functions and mechanisms of epigenetic inheritance in animals. Nat Rev Mol Cell Biol. 2018;19:774–90.
Talens RP, Christensen K, Putter H, Willemsen G, Christiansen L, Kremer D, et al. Epigenetic variation during the adult lifespan: cross-sectional and longitudinal data on monozygotic twin pairs. Aging Cell. 2012;11:694–703.
Goyal D, Limesand SW, Goyal R. Epigenetic responses and the developmental origins of health and disease. J Endocrinol. 2019;242:T105–19.
Lee HS. Impact of maternal diet on the epigenome during in utero life and the developmental programming of diseases in childhood and adulthood. Nutrients. 2015;7:9492–507.
Chango A, Pogribny IP. Considering maternal dietary modulators for epigenetic regulation and programming of the fetal epigenome. Nutrients. 2015;7:2748–70.
Lees C, Marlow N, Arabin B, Bilardo CM, Brezinka C, Derks JB, et al. TRUFFLE Group. Perinatal morbidity and mortality in early-onset fetal growth restriction: cohort outcomes of the trial of randomized umbilical and fetal flow in Europe (TRUFFLE). Ultrasound Obstet Gynecol. 2013;42:400–8.
Jaddoe VW, de Jonge LL, Hofman A, Franco OH, Steegers EA, Gaillard R. First trimester fetal growth restriction and cardiovascular risk factors in school age children: population based cohort study. BMJ. 2014;348: g14.
Miller SL, Huppi PS, Mallard C. The consequences of fetal growth restriction on brain structure and neurodevelopmental outcome. J Physiol. 2016;594:807–23.
Francis JH, Permezel M, Davey MA. Perinatal mortality by birthweight centile. Aust N Z J Obstet Gynaecol. 2014;54:354–9.
Baethge C, Goldbeck-Wood S, Mertens S. SANRA-a scale for the quality assessment of narrative review articles. Res Integr Peer Rev. 2019;4:5.
Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38.
Zhang ZM, Lu R, Wang P, Yu Y, Chen D, Gao L, et al. Structural basis for DNMT3A-mediated de novo DNA methylation. Nature. 2018;554:387–91.
Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA. 2006;103:1412–7.
Schulz WA, Steinhoff C, Florl AR. Methylation of endogenous human retroelements in health and disease. Curr Top Microbiol Immunol. 2006;310:211–50.
Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–82.
Lambertini L, Lee TL, Chan WY, Lee MJ, Diplas A, Wetmur J, Chen J. Differential methylation of imprinted genes in growth-restricted placentas. Reprod Sci. 2011;18:1111–7.
Gicquel C, El-Osta A, Le Bouc Y. Epigenetic regulation and fetal programming. Best Pract Res Clin Endocrinol Metab. 2008;22:1–16.
Wood AJ, Oakey RJ. Genomic imprinting in mammals: emerging themes and established theories. PLoS Genet. 2006;2: e147.
Nelissen EC, Van Montfoort AP, Dumoulin JC, Evers JL. Epigenetics and the placenta. Hum Reprod Update. 2011;17:397–417.
Wu WB, Xu YY, Cheng WW, Yuan B, Zhao JR, Wang YL, et al. Decreased PGF may contribute to trophoblast dysfunction in fetal growth restriction. Reproduction. 2017;154:319–29.
Randhawa R, Cohen P. The role of the insulin-like growth factor system in prenatal growth. Mol Genet Metab. 2005;86:84–90.
Nawathe AR, Christian M, Kim SH, Johnson M, Savvidou MD, Terzidou V. Insulin-like growth factor axis in pregnancies affected by fetal growth disorders. Clin Epigenet. 2016;8:11.
Börzsönyi B, Demendi C, Nagy Z, Tóth K, Csanád M, Pajor A, et al. Gene expression patterns of insulin-like growth factor 1, insulin-like growth factor 2 and insulin-like growth factor binding protein 3 in human placenta from pregnancies with intrauterine growth restriction. J Perinat Med. 2011;39:701–7.
Lee MH, Jeon YJ, Lee SM, Park MH, Jung SC, Kim YJ. Placental gene expression is related to glucose metabolism and fetal cord blood levels of insulin and insulin-like growth factors in intrauterine growth restriction. Early Hum Dev. 2010;86:45–50.
Xiao X, Zhao Y, Jin R, Chen J, Wang X, Baccarelli A, et al. Fetal growth restriction and methylation of growth-related genes in the placenta. Epigenomics. 2016;8:33–42.
Stewart PM, Murry BA, Mason JI. Type 2 11 beta-hydroxysteroid dehydrogenase in human fetal tissues. J Clin Endocrinol Metab. 1994;78:1529–32.
Marsit CJ, Maccani MA, Padbury JF, Lester BM. Placental 11-beta hydroxysteroid dehydrogenase methylation is associated with newborn growth and a measure of neurobehavioral outcome. PLoS ONE. 2012;7: e33794.
Zhao Y, Gong X, Chen L, et al. Site-specific methylation of placental HSD11B2 gene promoter is related to intrauterine growth restriction. Eur J Hum Genet. 2014;22:734–40.
Zhu P, Wang W, Zuo R, Sun K. Mechanisms for establishment of the placental glucocorticoid barrier, a guard for life. Cell Mol Life Sci. 2019;76:13–26.
Chelbi ST, Mondon F, Jammes H, Buffat C, Mignot TM, Tost J, et al. Expressional and epigenetic alterations of placental serine protease inhibitors: SERPINA3 is a potential marker of preeclampsia. Hypertension. 2007;49:76–83.
Chelbi ST, Wilson ML, Veillard AC, Ingles SA, Zhang J, Mondon F, et al. Genetic and epigenetic mechanisms collaborate to control SERPINA3 expression and its association with placental diseases. Hum Mol Genet. 2012;21:1968–78.
Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70.
Schaid DJ, Chen W, Larson NB. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat Rev Genet. 2018;19:491–504.
Hillman SL, Finer S, Smart MC, Mathews C, Lowe R, Rakyan VK, et al. Novel DNA methylation profiles associated with key gene regulation and transcription pathways in blood and placenta of growth-restricted neonates. Epigenetics. 2015;10:50–61.
Wutz A, Smrzka OW, Schweifer N, Schellander K, Wagner EF, Barlow DP. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745–9.
Wilhelm-Benartzi CS, Houseman EA, Maccani MA, Poage GM, Koestler DC, Langevin SM, et al. In utero exposures, infant growth, and DNA methylation of repetitive elements and developmentally related genes in human placenta. Environ Health Perspect. 2012;120:296–302.
Ferguson-Smith AC. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet. 2011;12:565–75.
McMinn J, Wei M, Schupf N, Cusmai J, Johnson EB, Smith AC, et al. Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta. 2006;27:540–9.
Tycko B. Imprinted genes in placental growth and obstetric disorders. Cytogenet Genome Res. 2006;113:271–8.
Tycko B, Morison IM. Physiological functions of imprinted genes. J Cell Physiol. 2002;192:245e58.
Higashimoto K, Jozaki K, Kosho T, Matsubara K, Fuke T, Yamada D, et al. A novel de novo point mutation of the OCT-binding site in the IGF2/H19-imprinting control region in a Beckwith-Wiedemann syndrome patient. Clin Genet. 2014;86:539–44.
Yamaguchi Y, Tayama C, Tomikawa J, Akaishi R, Kamura H, Matsuoka K, et al. Placenta-specific epimutation at H19-DMR among common pregnancy complications: its frequency and effect on the expression patterns of H19 and IGF2. Clin Epigenetics. 2019;11:113.
Tsunoda Y, Kudo M, Wada R, Ishino K, Kure S, Sakatani T, et al. Expression level of long noncoding RNA H19 of normotensive placentas in late pregnancy relates to the fetal growth restriction. J Obstet Gynaecol Res. 2020;46:1025–34.
Koukoura O, Sifakis S, Zaravinos A, Apostolidou S, Jones A, Hajiioannou J, et al. Hypomethylation along with increased H19 expression in placentas from pregnancies complicated with fetal growth restriction. Placenta. 2011;32:51–7.
Koukoura O, Sifakis S, Soufla G, Zaravinos A, Apostolidou S, Jones A, et al. Loss of imprinting and aberrant methylation of IGF2 in placentas from pregnancies complicated with fetal growth restriction. Int J Mol Med. 2011;28:481–7.
St-Pierre J, Hivert MF, Perron P, et al. IGF2 DNA methylation is a modulator of newborn’s fetal growth and development. Epigenetics. 2012;7:1125–32.
Bourque DK, Avila L, Penaherrera M, Von Dadelszen P, Robinson WP. Decreased placental methylation at the H19/IGF2 imprinting control region is associated with normotensive intrauterine growth restriction but not pre-eclampsia. Placenta. 2010;31:197–202.
Cordeiro A, Neto AP, Carvalho F, Ramalho C, Doria S. Relevance of genomic imprinting in intrauterine human growth expression of CDKN1C, H19, IGF2, KCNQ1 and PHLDA2 imprinted genes. J Assist Reprod Genet. 2014;31:1361–8.
Stalman SE, Solanky N, Ishida M, Alemán-Charlet C, Abu-Amero S, Alders M, et al. Genetic analyses in small-for-gestational-age newborns. J Clin Endocrinol Metab. 2018;103:917–25.
Eggermann T, Binder G, Brioude F, Maher ER, Lapunzina P, Cubellis MV, et al. CDKN1C mutations: two sides of the same coin. Trends Mol Med. 2014;20:614–22.
Caniçais C, Vasconcelos S, Ramalho C, Marques CJ, Dória S. Deregulation of imprinted genes expression and epigenetic regulators in placental tissue from intrauterine growth restriction. J Assist Reprod Genet. 2021;38:791–801.
Diplas AI, Lambertini L, Lee MJ, Sperling R, Lee YL, Wetmur J, et al. Differential expression of imprinted genes in normal and IUGR human placentas. Epigenetics. 2009;4:235–40.
Zhao Z, Tavoosidana G, Sjölinder M, Göndör A, Mariano P, Wang S, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–7.
Sutton VR, Shaffer LG. Search for imprinted regions on chromosome 14: comparison of maternal and paternal UPD cases with cases of chromosome 14 deletion. Am J Med Genet. 2000;93:381–7.
Wylie AA, Murphy SK, Orton TC, Jirtle RL. Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation. Genome Res. 2000;10:1711–8.
Sekita Y, Wagatsuma H, Nakamura K, Ono R, Kagami M, Wakisaka N, et al. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat Genet. 2008;40:243e8.
Fujioka K, Nishida K, Ashina M, Abe S, Fukushima S, Ikuta T, Ohyama S, Morioka I, Iijima K. DNA methylation of the Rtl1 promoter in the placentas with fetal growth restriction. Pediatr Neonatol. 2019;60:512–6.
Barlow DP, Bartolomei MS. Genomic imprinting in mammals. Cold Spring Harb Perspect Biol. 2014;6: a018382.
Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–95.
Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci USA. 1964;51:786–94.
Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–32.
Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80.
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74.
Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–8.
Alsat E, Guibourdenche J, Couturier A, Evain-Brion D. Physiological role of human placental growth hormone. Mol Cell Endocrinol. 1998;140:121–7.
Kimura AP, Liebhaber SA, Cooke NE. Epigenetic modifications at the human growth hormone locus predict distinct roles for histone acetylation and methylation in placental gene activation. Mol Endocrinol. 2004;18:1018–32.
Paauw ND, Lely AT, Joles JA, Franx A, Nikkels PG, Mokry M, et al. H3K27 acetylation and gene expression analysis reveals differences in placental chromatin activity in fetal growth restriction. Clin Epigenet. 2018;10:85.
Maltepe E, Krampitz GW, Okazaki KM, Red-Horse K, Mak W, Simon MC, et al. Hypoxia-inducible factor-dependent histone deacetylase activity determines stem cell fate in the placenta. Development. 2005;132:3393–403.
Alahari S, Post M, Rolfo A, Weksberg R, Caniggia I. Compromised JMJD6 histone demethylase activity affects VHL gene repression in preeclampsia. J Clin Endocrinol Metab. 2018;103:1545–57.
Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40.
Gebremedhn S, Ali A, Hossain M, Hoelker M, Salilew-Wondim D, Anthony RV, et al. MicroRNA-mediated gene regulatory mechanisms in mammalian female reproductive health. Int J Mol Sci. 2021;22:938.
Frías-Lasserre D, Villagra CA. The Importance of ncRNAs as epigenetic mechanisms in phenotypic variation and organic evolution. Front Microbiol. 2017;8:2483.
Wei JW, Huang K, Yang C, Kang CS. Non-coding RNAs as regulators in epigenetics (Review). Oncol Rep. 2017;37:3–9.
Chisholm KM, Wan Y, Li R, Montgomery KD, Chang HY, West RB. Detection of long non-coding RNA in archival tissue: correlation with polycomb protein expression in primary and metastatic breast carcinoma. PLoS ONE. 2012;7: e47998.
Martinez VD, Cohn DE, Telkar N, Minatel BC, Pewarchuk ME, Marshall EA, et al. Profiling the small non-coding RNA transcriptome of the human placenta. Sci Data. 2021;8:166.
Santoro G, Lapucci C, Giannoccaro M, Caporilli S, Rusin M, Seidenari A, et al. Abnormal circulating maternal miRNA expression is associated with a low (< 4%) cell-free DNA fetal fraction. Diagn (Basel). 2021;11:2108.
Addo KA, Palakodety N, Hartwell HJ, Tingare A, Fry RC. Placenta microRNAs: responders to environmental chemicals and mediators of pathophysiology of the human placenta. Toxicol Rep. 2020;7:1046–56.
Hromadnikova I. Extracellular nucleic acids in maternal circulation as potential biomarkers for placental insufficiency. DNA Cell Biol. 2012;31:1221–32.
Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–70.
Xie L, Mouillet JF, Chu T, Parks WT, Sadovsky E, Knöfler M, et al. C19MC microRNAs regulate the migration of human trophoblasts. Endocrinology. 2014;155:4975–85.
Mong EF, Yang Y, Akat KM, Canfield J, VanWye J, Lockhart J, et al. Chromosome 19 microRNA cluster enhances cell reprogramming by inhibiting epithelial-to-mesenchymal transition. Sci Rep. 2020;10:3029.
Morales-Prieto DM, Ospina-Prieto S, Chaiwangyen W, Schoenleben M, Markert UR. Pregnancy-associated miRNA-clusters. J Reprod Immunol. 2013;97:51–61.
Gu Y, Sun J, Groome LJ, Wang Y. Differential miRNA expression profiles between the first and third trimester human placentas. Am J Physiol Endocrinol Metab. 2013;304:E836–43.
Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genom. 2007;8:166.
Kochhar P, Vukku M, Rajashekhar R, Mukhopadhyay A. microRNA signatures associated with fetal growth restriction: a systematic review. Eur J Clin Nutr. 2022;76:1088–102.
Ghosh G, Subramanian IV, Adhikari N, Zhang X, Joshi HP, Basi D, et al. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-α isoforms and promotes angiogenesis. J Clin Investig. 2010;120:4141–54.
Fasanaro P, D’Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–83.
Zou Z, Forbes K, Harris LK, Heazell AEP. The potential role of the E SRRG pathway in placental dysfunction. Reproduction. 2021;161:R45-60.
Takeda Y, Liu X, Sumiyoshi M, Matsushima A, Shimohigashi M, Shimohigashi Y. Placenta expressing the greatest quantity of bisphenol A receptor ERR{gamma} among the human reproductive tissues: predominant expression of type-1 ERRgamma isoform. J Biochem. 2009;146:113–22.
Zhu H, Huang L, He Z, Zou Z, Luo Y. Estrogen-related receptor γ regulates expression of 17β-hydroxysteroid dehydrogenase type 1 in fetal growth restriction. Placenta. 2018;67:38–44.
Backes C, Meese E, Keller A. Specific miRNA disease biomarkers in blood, serum and plasma: challenges and prospects. Mol Diagn Ther. 2016;20:509–18.
Kotlabova K, Doucha J, Hromadnikova I. Placental-specific microRNA in maternal circulation: identification of appropriate pregnancy-associated microRNAs with diagnostic potential. J Reprod Immunol. 2011;89:185–91.
Luo X, Li X. Long non-coding RNAs serve as diagnostic biomarkers of preeclampsia and modulate migration and invasiveness of trophoblast cells. Med Sci Monit. 2018;24:84–91.
Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10:28–36.
Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–12.
Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313–6.
Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–65.
Koerner MV, Pauler FM, Huang R, Barlow DP. The function of non-coding RNAs in genomic imprinting. Development. 2009;136:1771–83.
Gremlich S, Damnon F, Reymondin D, Braissant O, Schittny JC, Baud D, et al. The long non-coding RNA NEAT1 is increased in IUGR placentas, leading to potential new hypotheses of IUGR origin/development. Placenta. 2014;35:44–9.
Huang X, Anderle P, Hostettler L, Baumann MU, Surbek DV, Ontsouka EC, et al. Identification of placental nutrient transporters associated with intrauterine growth restriction and pre-eclampsia. BMC Genom. 2018;19:173.
Monteiro LJ, Peñailillo R, Sánchez M, Acuña-Gallardo S, Mönckeberg M, Ong J, et al. The role of long non-coding RNAs in trophoblast regulation in preeclampsia and intrauterine growth restriction. Genes (Basel). 2021;12:970.
Medina-Bastidas D, Guzmán-Huerta M, Borboa-Olivares H, Ruiz-Cruz C, Parra-Hernández S, Flores-Pliego A, et al. Placental microarray profiling reveals common mRNA and lncRNA expression patterns in preeclampsia and intrauterine growth restriction. Int J Mol Sci. 2020;21:3597.
Lipka A, Jastrzebski JP, Paukszto L, Makowczenko KG, Lopienska-Biernat E, Gowkielewicz M, et al. Sex-biased lncRNA signature in fetal growth restriction (FGR). Cells. 2021;10:921.
Hitchins MP. Inheritance of epigenetic aberrations (constitutional epimutations) in cancer susceptibility. Adv Genet. 2010;70:201–43.
Terstappen F, Calis JJA, Paauw ND, Joles JA, van Rijn BB, Mokry M, et al. Developmental programming in human umbilical cord vein endothelial cells following fetal growth restriction. Clin Epigenet. 2020;12:185.
Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat Rev Genet. 2012;13:153–62.
Xavier MJ, Roman SD, Aitken RJ, Nixon B. Transgenerational inheritance: how impacts to the epigenetic and genetic information of parents affect offspring health. Hum Reprod Update. 2019;25:518–40.
Roberts CT. IFPA Award in Placentology Lecture: complicated interactions between genes and the environment in placentation, pregnancy outcome and long term health. Placenta. 2010;Suppl.:S47–53.
Mikheev AM, Nabekura T, Kaddoumi A, Bammler TK, Govindarajan R, Hebert MF, et al. Profiling gene expression in human placentae of different gestational ages: an OPRU Network and UW SCOR Study. Reprod Sci. 2008;15:866–77.
Yuen RK, Avila L, Peñaherrera MS, von Dadelszen P, Lefebvre L, Kobor MS, et al. Human placental-specific epipolymorphism and its association with adverse pregnancy outcomes. PLoS ONE. 2009;4: e7389.
Novakovic B, Yuen RK, Gordon L, Penaherrera MS, Sharkey A, Moffett A, et al. Evidence for widespread changes in promoter methylation profile in human placenta in response to increasing gestational age and environmental/stochastic factors. BMC Genom. 2011;12:529.
Zdravkovic T, Genbacev O, McMaster MT, Fisher SJ. The adverse effects of maternal smoking on the human placenta: a review. Placenta. 2005;26(Suppl. A):S81–6.
Chandravanshi L, Shiv K, Kumar S. Developmental toxicity of cadmium in infants and children: a review. Environ Anal Health Toxicol. 2021;36: e2021003.
Lee S, Hong YC, Park H, Kim Y, Ha M, Ha E. Combined effects of multiple prenatal exposure to pollutants on birth weight: the Mothers and Children’s Environmental Health (MOCEH) study. Environ Res. 2020;181: 108832.
Rolfo A, Nuzzo AM, De Amicis R, Moretti L, Bertoli S, Leone A. Fetal-maternal exposure to endocrine disruptors: correlation with diet intake and pregnancy outcomes. Nutrients. 2020;12:1744.
Joubert BR, Håberg SE, Bell DA, Nilsen RM, Vollset SE, Midttun O, et al. Maternal smoking and DNA methylation in newborns: in utero effect or epigenetic inheritance? Cancer Epidemiol Biomark Prev. 2014;23:1007–17.
Flom JD, Ferris JS, Liao Y, Tehranifar P, Richards CB, Cho YH, et al. Prenatal smoke exposure and genomic DNA methylation in a multiethnic birth cohort. Cancer Epidemiol Biomark Prev. 2011;20:2518–23.
Joubert BR, Haberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect. 2012;120:1425–31.
Ghazi T, Naidoo P, Naidoo RN, Chuturgoon AA. Prenatal air pollution exposure and placental DNA methylation changes: implications on fetal development and future disease susceptibility. Cells. 2021;10:3025.
Zhao Y, Shi HJ, Xie CM, Chen J, Laue H, Zhang YH. Prenatal phthalate exposure, infant growth, and global DNA methylation of human placenta. Environ Mol Mutagen. 2015;56:286–92.
Zhao Y, Song Q, Ge W, Jin Y, Chen S, Zhao Y, et al Y. Associations between in utero exposure to polybrominated diphenyl ethers, pathophysiological state of fetal growth and placental DNA methylation changes. Environ Int. 2019;133(Pt B):105255.
Li LX, Chen L, Meng XZ, Chen BH, Chen SQ, Zhao Y, et al. Exposure levels of environmental endocrine disruptors in mother-newborn pairs in China and their placental transfer characteristics. PLoS ONE. 2013;8: e62526.
Susiarjo M, Sasson I, Mesaros C, Bartolomei MS. Bisphenol a exposure disrupts genomic imprinting in the mouse. PLoS Genet. 2013;9: e1003401.
Wang B, Liu JJ, Wang Y, Fu L, Shen R, Yu Z, et al. Maternal fenvalerate exposure induces fetal intrauterine growth restriction through disrupting placental thyroid hormone receptor signaling. Toxicol Sci. 2017;157:377–86.
Yu Z, Han Y, Shen R, Huang K, Xu YY, Wang QN, et al. Gestational di-(2-ethylhexyl) phthalate exposure causes fetal intrauterine growth restriction through disturbing placental thyroid hormone receptor signaling. Toxicol Lett. 2018;294:1–10.
Chen CY, Chen CP, Lin KH. Biological functions of thyroid hormone in placenta. Int J Mol Sci. 2015;16:4161–79.
O’Callaghan JL, Clifton VL, Prentis P, Ewing A, Miller YD, Pelzer ES. Modulation of placental gene expression in small-for-gestational-age infants. Genes (Basel). 2020;11:80.
O’Callaghan JL, Turner R, Dekker Nitert M, Barrett HL, Clifton V, Pelzer ES. Re-assessing microbiomes in the low-biomass reproductive niche. BJOG. 2020;127:147–58.
Parnell LA, Briggs CM, Cao B, Delannoy-Bruno O, Schrieffer AE, Mysorekar IU. Microbial communities in placentas from term normal pregnancy exhibit spatially variable profiles. Sci Rep. 2017;7:11200.
Barak S, Oettinger-Barak O, Machtei EE, Sprecher H, Ohel G. Evidence of periopathogenic microorganisms in placentas of women with preeclampsia. J Periodontol. 2007;78:670–6.
Doyle RM, Harris K, Kamiza S, Harjunmaa U, Ashorn U, Nkhoma M, et al. Bacterial communities found in placental tissues are associated with severe chorioamnionitis and adverse birth outcomes. PLoS ONE. 2017;12: e0180167.
Zheng J, Xiao X, Zhang Q, Mao L, Yu M, Xu J. The placental microbiome varies in association with low birth weight in full-term neonates. Nutrients. 2015;7:6924–37.
Viuff AC, Pedersen LH, Kyng K, Staunstrup NH, Børglum A, Henriksen TB. Antidepressant medication during pregnancy and epigenetic changes in umbilical cord blood: a systematic review. Clin Epigenet. 2016;8:94.
Argyraki M, Damdimopoulou P, Chatzimeletiou K, Grimbizis GF, Tarlatzis BC, Syrrou M, et al. In-utero stress and mode of conception: impact on regulation of imprinted genes, fetal development and future health. Hum Reprod Update. 2019;25:777–801.
Rhon-Calderon EA, Vrooman LA, Riesche L, Bartolomei MS. The effects of assisted reproductive technologies on genomic imprinting in the placenta. Placenta. 2019;84:37–43.
Lazaraviciute G, Kauser M, Bhattacharya S, Haggarty P, Bhattacharya S. A systematic review and meta-analysis of DNA methylation levels and imprinting disorders in children conceived by IVF/ICSI compared with children conceived spontaneously. Hum Reprod Update. 2014;20:840–52.
El Hajj N, Haaf T. Epigenetic disturbances in in vitro cultured gametes and embryos: implications for human assisted reproduction. Fertil Steril. 2013;99:632–41.
Zhu HL, Shi XT, Xu XF, Zhou GX, Xiong YW, Yi SJ, et al. Melatonin protects against environmental stress-induced fetal growth restriction via suppressing ROS-mediated GCN2/ATF4/BNIP3-dependent mitophagy in placental trophoblasts. Redox Biol. 2021;40: 101854.
Zhang L, Lu Q, Chang C. Epigenetics in health and disease. Adv Exp Med Biol. 2020;1253:3–55.
Chatterjee S, Ouidir M, Tekola-Ayele F. Genetic and in utero environmental contributions to DNA methylation variation in placenta. Hum Mol Genet. 2021;30:1968–76.
Abdulghani M, Jain A, Tuteja G. Genome-wide identification of enhancer elements in the placenta. Placenta. 2019;79:72–7.
Smith E, Shilatifard A. Enhancer biology and enhanceropathies. Nat Struct Mol Biol. 2014;21:210–9.
Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science. 1990;248:220–3.
Ferreira LMR, Meissner TB, Tilburgs T, Strominger JL. HLA-G: at the interface of maternal fetal tolerance. Trends Immunol. 2017;38:272–86.
Kikas T, Laan M, Kasak L. Current knowledge on genetic variants shaping placental transcriptome and their link to gestational and postnatal health. Placenta. 2021;116:2–11.
Zanello M, DeSanctis P, Pula G, Zucchini C, Pittalis MC, Rizzo N, et al. Circulating mRNA for epidermal growth factor-like domain 7 (EGFL7) in maternal blood and early intrauterine growth restriction: a preliminary analysis. Prenat Diagn. 2013;33:168–72.
Whitehead CL, Walker SP, Mendis S, Lappas M, Tong S. Quantifying mRNA coding growth genes in the maternal circulation to detect fetal growth restriction. Am J Obstet Gynecol. 2013;209(133):e1-9.
Murthi P. Review: placental homeobox genes and their role in regulating human fetal growth. Placenta. 2014;35(Suppl.):S46–50
Sharma D, Sharma P, Shastri S. Genetic, metabolic and endocrine aspect of intrauterine growth restriction: an update. J Matern Fetal Neonatal Med. 2017;30:2263–75.
Majewska M, Lipka A, Paukszto L, Jastrzebski JP, Szeszko K, Gowkielewicz M, et al. Placenta transcriptome profiling in intrauterine growth restriction (IUGR). Int J Mol Sci. 2019;20(6):1510.
Murthi P, Said JM, Doherty VL, Donath S, Nowell CJ, Brennecke SP, et al. Homeobox gene DLX4 expression is increased in idiopathic human fetal growth restriction. Mol Hum Reprod. 2006;12:763–9.
Gascoin-Lachambre G, Buffat C, Rebourcet R, Chelbi ST, Rigourd V, Mondon F, et al. Cullins in human intra-uterine growth restriction: expressional and epigenetic alterations. Placenta. 2010;31:151–7.
Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. Genome Biol. 2011;12:220.
Gong S, Gaccioli F, Dopierala J, Sovio U, Cook E, Volders PJ, et al. The RNA landscape of the human placenta in health and disease. Nat Commun. 2021;12:2639.
Jin XX, Ying X, Dong MY. Galectin-1 expression in the serum and placenta of pregnant women with fetal growth restriction and its significance. BMC Pregnancy Childbirth. 2021;21:14.
Chabrun F, Huetz N, Dieu X, Rousseau G, Bouzillé G, Chao de la Barca JM, et al. Data-mining approach on transcriptomics and methylomics placental analysis highlights genes in fetal growth restriction. Front Genet. 2020;10:1292.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
No funding was received for the preparation of this article.
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Code availability
Not applicable.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Salmeri, N., Carbone, I., Cavoretto, P. et al. Epigenetics Beyond Fetal Growth Restriction: A Comprehensive Overview. Mol Diagn Ther 26, 607–626 (2022). https://doi.org/10.1007/s40291-022-00611-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-022-00611-4