Abstract
Introduction
Comprehensive genetic cancer profiling using circulating tumor DNA has enabled the detection of National Comprehensive Cancer Network (NCCN) guideline-recommended somatic alterations from a single, non-invasive blood draw. However, reliably detecting somatic variants at low variant allele fractions (VAFs) remains a challenge for next-generation sequencing (NGS)-based tests. We have developed the single-molecule sequencing (SMSEQ) platform to address these challenges.
Methods
The OncoLBx assay utilizes the SMSEQ platform to optimize cell-free DNA extraction and library preparation with variant type-specific calling algorithms to improve sensitivity and specificity. OncoLBx is a pan-cancer panel for solid tumors targeting 75 genes and five microsatellite sites analyzing five classes of NCCN-recommended somatic variants: single-nucleotide variants (SNVs), insertions and deletions (indels), copy number variants (CNVs), fusions and microsatellite instability (MSI). Circulating DNA was extracted from plasma, followed by library preparation using SMSEQ. Analytical validation was performed according to recently published American College of Medical Genetics and Genomics (ACMG)/Association for Molecular Pathology (AMP) guidelines and established the limit of detection (LOD), sensitivity, specificity, accuracy and reproducibility using 126 gold-standard reference samples, healthy donor samples verified by whole-exome sequencing by an external College of American Pathologists (CAP) reference lab and cell lines with known variants. Results were analyzed using a locus-specific modeling algorithm.
Results
We have demonstrated that OncoLBx detects VAFs of ≥ 0.1% for SNVs and indels, ≥ 0.5% for fusions, ≥ 4.5 copies for CNVs and ≥ 2% for MSI, with all variant types having specificity ≥ 99.999%. Diagnostic performance of paired samples displays 80% sensitivity and > 99.999% clinical specificity. Clinical utility and performance were assessed in 416 solid tumor samples. Variants were detected in 79% of samples, for which 87.34% of positive samples had available targeted therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
This article outlines the creation and rigorous validation of a novel single-molecule sequencing (SMSEQ) platform and a custom gene panel covering 75 genes and five microsatellite site markers for diagnosis of cancer and therapy selection. |
We assay National Comprehensive Cancer Network (NCCN) guideline-recommended somatic genomic alterations for solid tumors using a single, non-invasive blood draw. |
The platform analyzes, and we report the validation of, five classes of somatic variants: single-nucleotide variants, insertions and deletions, copy number variants, fusions, and microsatellite instability. |
1 Introduction
The increased use of next-generation sequencing (NGS) in laboratory-developed tests (LDTs) has facilitated the transition of this technology to clinical application for cancer genomic profiling [1]. Genetic analysis of tumors has driven the development and clinical use of targeted and immunotherapy for cancer treatment. For example, trastuzumab (Herceptin) is a targeted therapy with well-demonstrated efficacy in breast cancer and pembrolizumab, a checkpoint immunotherapy agent, was recently approved by the Food and Drug Administration (FDA) as a pan-tumor indication for patients with cancers harboring high microsatellite instability (MSI-H) or mismatch repair deficiency (dMMR) [2,3,4,5,6]. NGS based tests allow simultaneous testing of thousands of loci and parallel analysis of different classes of genetic variants in a single assay. In contrast, other methods such as digital polymerase chain reaction (dPCR) may require sequential or multiple tests, or excessive material to cover the breadth and depth that NGS-based assays can achieve from a single assay [1, 7]. For example, BRAF V600E-mutated cancers, rare in non-small cell lung cancer, are not routinely tested for and are generally only identified through broad test panels, often from NGS platforms [8,9,10]. As new therapies, genetic targets, and target types emerge, LDTs are poised to be the first to translate new discoveries into validated clinical tests, with NGS being one of the most common platforms for new test development [1, 7].
NGS-based tumor tissue biopsy tests have demonstrated clinical utility for therapy selection [11, 12]. Tissue biopsies, when possible to obtain, are limited in scope to small specimens. Many cancer types are genetically heterogeneous, and tumor tissue obtained by tissue biopsy may not fully reflect this heterogeneity, resulting in false negative results [3, 13]. Further, tissue biopsy, as an invasive diagnostic test, can be risky and has relatively low levels of patient acceptance [14]. Moreover, for patients in late metastatic stages of cancer, acquired mutations from continued tumor evolution at both primary and metastatic sites may lead to misleading results [15,16,17]. These shortcomings have fueled research and development of liquid biopsy as an alternative to tissue biopsy.
Physiological events of cells such as apoptosis, necrosis and secretion release both cell-free DNA (cfDNA) from normal tissue as well as tumor-derived cfDNA (ctDNA) into the bloodstream [18]. It has been shown that variants of tumor origin can be identified in ctDNA [18,19,20]. Because cfDNA is released via physiological events, it might have a more global genomic view across the heterogeneous landscape of primary tumors and their metastases. In addition, cfDNA has a short half-life (~ 2 h), which means that blood can be used as a “real-time” reflection of the genetics of the tumor at the time of blood draw [21,22,23,24,25].
However, there are major challenges that limit the use of standard NGS techniques and protocols for accurate and reproducible detection of ctDNA in blood. Generally, there is a limited amount of cfDNA fragments present in peripheral blood, and within the cfDNA, only a small fraction of molecules may be of tumor origin (ctDNA) [26]. The tumor fraction of cfDNA, which varies by cancer type, stage and location, has been reported at low variant allele fractions (VAFs) (0.1%) [27,28,29,30]. At such low VAFs, the intrinsic error rate of standard NGS renders it difficult to distinguish tumor-derived variants from various sources of error that occur during library preparation, cluster formation, and sequencing [30,31,32]. To overcome these technical hurdles, we have developed a novel platform, single-molecule sequencing (SMSEQ), that is optimized for isolating and analyzing ctDNA to identify five classes of variants at low allele frequencies.
In this study, we paired the SMSEQ platform with a pan-cancer NGS panel targeting 75 genes and 5 microsatellite sites to create a liquid biopsy test, OncoLBx. This panel targets five types of variants (single-nucleotide variants [SNVs], insertions and deletions [indels], copy number variants [CNVs], fusions and microsatellite instability [MSI]) spanning all National Comprehensive Cancer Network (NCCN) guideline-recommended actionable genomic variants for solid tumors (see the electronic supplementary material [ESM] methods for the list of genes targeted for each variant type). We have performed validation and benchmarking of OncoLBx following the recently published College of American Pathologists (CAP)/Association for Molecular Pathology (AMP) guidelines for the development and validation of NGS-based oncology panels [33]. OncoLBx was validated against a wide range of sample types, including gold-standard reference samples, cell lines with known variants, blood and tissue pairs, and clinical samples whose variants have been verified by an external Clinical Laboratory Improvement Amendments (CLIA)/CAP lab. In addition, we show clinical results from our first 416 consecutive samples sent to our CAP-accredited laboratory in Taipei, Taiwan.
2 Materials and Methods
2.1 Analytical Samples
Analytical control samples and cell lines were acquired from commercial vendors (see supplementary Table S3 in the ESM). Samples used for analytical validation were derived from commercial reference samples designed and verified to contain known variants at specific allele frequencies or cell lines known to harbor specific variants (see the supplementary methods for details).
2.2 Clinical Samples
Institutional review board (IRB) and informed consent was obtained from Chang Gung Memorial Hospital for 97 study patients, including 36 patients providing both blood and tissue. Informed consent was obtained from all 97 study patients and for 319 consecutive clinical samples that were processed by CellMax Life’s CAP-accredited lab in Taiwan and a CLIA-certified and CAP-accredited US laboratory. All patient information presented has been de-identified and anonymized.
2.3 Collection of Blood and Isolation of Plasma and Buffy Coat
Ten milliliters of blood were collected from patients and healthy donors in Cell-Free DNA BCT (Streck) and K3/EDTA tubes. The tube was spun down at 1600 g for 10 min at room temperature to separate plasma, buffy coat and red blood cells. The plasma fraction was carefully isolated and further spun down at 16,000 g for 10 min to collect cell-free plasma.
2.4 Extraction of cfDNA
Cell-free DNA (cfDNA) was extracted from approximately 4–5 ml of plasma using the Qiagen Circulating Nucleic Acid kit with an optimized protocol to increase cfDNA extraction efficiency. Incubation time for proteinase K and lysis treatment was extended twofold from standard protocol to maximize cfDNA release. In addition, carrier RNA was used to minimize cfDNA loss during the extraction process. To further enhance the cfDNA recovery rate, double elution was performed using eluate from first elution for the second elution, with an incubation time of 15 min prior to elution (standard protocol is single elution and 3-min incubation time). See the supplementary methods in the ESM for cfDNA quantification and quality control (QC).
2.5 SMSEQ Library preparation
Libraries were prepared using 20–30 ng size corrected input cfDNA (see the supplementary methods in the ESM). The ends of the cfDNA were repaired (blunt-ended and 5′ phosphorylated) enzymatically and dA-tailed (see the supplementary methods). In order to perform SMSEQ analysis, unique molecular indices (UMIs) were ligated on one end and Tru-Seq single-index sequencing adapters from Integrated DNA Technologies (IDT) were ligated on the other. Both adapters include sequences complementary to Illumina’s flow-cell sequence to facilitate sequence reading (see the supplementary methods for SMSEQ adapter design). In order to achieve maximum ligation efficiency, we lowered the standard ligation temperature to 18 °C and extended the ligation time to 4 h. In order to reduce polymerase errors and amplification biases, the ligated cfDNA libraries were polymerase chain reaction (PCR) amplified with a minimum number of cycles (five cycles) using high-fidelity, low-error polymerase using primers directed against common ends. This was followed by double-sided Ampure XP (Beckman Coulter) size selection and cleanup to generate sequencing libraries with sample-specific unique barcodes. See the supplementary methods for workflow and QC.
2.6 Panel Design
SMSEQ target enrichment was performed with a custom 75-gene capture panel covering SNVs, indels, CNVs, fusions and MSI across 85 kb bases. The panel consists of approximately 700 biotinylated DNA probes which are 120 bp long. The DNA probes were designed by CellMax and manufactured by IDT. The probes were pooled together to constitute a panel.
2.7 Hybridization/Target Enrichment
A custom proprietary hybridization buffer was created specifically for the custom panel with ionic strength and additives based on the capture probe complexity, GC content, and probe length. Hybridization/wash time and temperature were carefully tuned based on the dynamic range of the probe’s Tm. With extensive optimization of this step, we were able to decrease the off-target rate and minimize GC bias by performing capture at a temperature of 62 °C for 4 h. This step was followed by a pull-down using streptavidin beads (see the supplementary methods for details).
2.8 Sequencing
The enriched libraries (capture pools) are quantified by quantitative PCR and normalized and pooled together to ensure adequate sequence coverage. 0.5-nM libraries are denatured with freshly made 0.2 N NaOH and diluted down to 21 pM. PhiX is spiked in, and the final pool is run in Illumina MiSeq using a 600-cycle V3 kit. Paired-end read lengths (> 200 bp) are used to ensure that there is significant overlap between the forward and reverse reads on average for each library strand, to increase accuracy. See the supplementary methods for QC details.
2.9 Informatics Pipeline and Sequencing Data Processing
OncoLBx sequencing data are processed by a custom bioinformatics pipeline built on common, established open-source tools, with software provided by Illumina and proprietary software designed and optimized for use with SMSEQ. See the supplementary methods for details.
2.10 Primary and Secondary Analysis
Primary and secondary analysis are performed using tools provided by Illumina and other open-source software to produce BAM files aligned to the human genome. Alignments are then used by SMSEQ for variant calling. See supplementary methods for details.
2.11 SMSEQ Analysis
Custom software was developed to utilize UMIs ligated to each cfDNA molecule (parent molecule) during library preparation. Daughter molecules created from parent molecules during amplification are grouped by UMI and alignment position. Positions with quality scores < Q30 are ignored, and low-quality tails of reads are trimmed. In addition, daughter molecules with lengths < 100 bases are filtered out. After filtering, reads from both forward and reverse strands are used to build a consensus sequence. For each position, the base with ≥ 75% consensus among all daughter molecules is selected as the consensus base and non-consensus bases are treated as errors and ignored. This consensus criteria results in the removal of errors (false positives).
2.11.1 SMSEQ SNV and Short Indel Analysis
SMSEQ’s SNVs and indel variant calling is performed by consensus read analysis across all target regions in the 75-gene panel. SMSEQ analyzes every position by using SMSEQ data and comparing potential variants against a position-specific scoring matrix (PSSM), to separate true variants from background noise (Fig. 1). Variants are evaluated using the following equation:
where RX is the sequencing data for sample X, a is the alternate allele being evaluated, L is the genomic location of the variant, M is the PSSM for a specific locus (L), G is the germline genotype of sample X at location L, and the significance threshold, α, is defined for each locus. If P(R|M) < α, then the variant is a call candidate. P(R|M) is distributed as a beta distribution with parameters derived from the PSSM. Candidate variants are evaluated for single-molecule support and other sample specific-factors such as background signal.
Single-nucleotide variants detection performance using single-molecule sequencing (SMSEQ). Graphs a and b show an example of the results of SMSEQ analysis on cell-line data with known variants. False positives are colored in black, and expected true positives are red. The red dashed lines represent the calling region for the sample. The line at 0.1% is for the lower limit of detection. a The results of the data when no SMSEQ error correction analysis is performed. b The results after performing SMSEQ error correction on the same data set. c The distribution of KRAS G12R allele frequencies was plotted for all 416 clinical samples. The red dotted line represents the statistical significance allele frequency cutoff (p value cutoff) and the blue line represents the coverage cutoff (allele frequency cutoff). The dashed black line shows the position-specific scoring matrix (PSSM)-based statistical distribution. Samples passing all cutoff thresholds are called positive and are shown shaded in black. d An example of KRAS G12R p values generated for variant calling in OncoLBx. Samples called positive are marked in red. Positive samples show significant separation from negatives
2.11.2 SMSEQ Copy Number Analysis
CNVs are determined by performing coverage analysis across all targets in the OncoLBx panel at both the gene and exon level. Coverage for all targets is normalized and compared against genomic DNA (gDNA) to create a sample-specific copy number profile. This profile is compared against similar profiles from healthy samples to identify copy number variations. These candidates are compared against expected values to determine if a CNV is present. CNVs are viewed in the context of the copy numbers of nearby genomic markers and observed allelic imbalance.
2.11.3 SMSEQ Gene Fusion Detection
Fusions candidates are identified by assessing their predicted breakpoints and single-molecule support. Gene fusion evaluation is performed targeting introns of five genes (FGFR3, NTRK1, RET, ROS1, and ALK) with canonical fusion events and established treatment indications based on NCCN guidelines. Reads which pass QC are tested for breakpoints by split read analysis and filtered for target regions. Split reads are divided into fragments based on their alignments. Read fragments are realigned to the genome and recombined together to determine the breakpoint. Potential breakpoints are evaluated by their level of single-molecule support and checked against expected background noise, with those passing considered fusion candidates. Candidates are then evaluated by their predicted protein and functional consequences, and known treatment indications.
2.11.4 SMSEQ MSI Status Determination
MSI status is determined by tabulating homopolymer length changes at five well-established microsatellite sites: NR-21, NR-24, BAT-25, BAT-26 and MONO-27 [34, 35]. Homopolymer length is determined by realigning raw reads to MSI target regions. The lengths of homopolymers observed are converted into a homopolymer score. The homopolymer score is compared against normal samples to generate a p value. This p value is used to determine whether a specific site is microsatellite stable (MSS) or microsatellite unstable. MSI status is determined by assessing these sites. Samples with two or more unstable sites are considered as MSI-H, one unstable site as MSI-low, and 0 unstable sites as MSS.
2.12 Calculation and Definition of Performance Metrics
Analytical performance of OncoLBx was established by comparing the set of called variants against the set of expected known variants according to the guidelines published by the AMP/CAP for designing clinical oncology LDTs using NGS [33] (see Sect. 2). The analytical validation includes 126 samples derived from 19 established cell lines with droplet digital PCR (ddPCR)-verified variants, ten healthy donors verified by exome sequencing from an external CAP/CLIA lab, and cfDNA reference control samples. A variant is considered negative if there is no verified variant at that target. For indels and SNVs, every locus in the panel is considered a target. For other variant types, a negative result simply is defined as no variant detected for that target. Diagnostic performance was assessed by comparing call sets against one another (see the supplementary methods for details).
3 Results
3.1 SMSEQ Analysis
SMSEQ analysis utilizes UMIs to suppress errors and to improve variant calling accuracy (see Sect. 2). A cancer cell line with known somatic variants (true positives) was used to evaluate the performance of SMSEQ error correction across a large genomic region. As shown in Fig. 1a, analysis without the error correction shows a large number of false positives at lower allele frequencies (most under 0.2%). It is not possible to separate true positives (red) from false positives (black). The error correction removes false positives, while maintaining the signal from true positives (red) at expected allele frequencies (Fig. 1b). These results demonstrate that SMSEQ error correction facilitates the separation of true and false positives, which improves the limit of detection (LOD) and accuracy of OncoLBx.
3.2 SNV Detection Performance
SNVs are detected using SMSEQ and custom algorithms trained to evaluate and separate variants from background. SMSEQ variant calling utilizes a PSSM which has been developed and trained against a separate set of 125 samples from healthy donors, patient samples, analytical controls, and cell lines to separate variants from background (see Sect. 2). P(R|M) is modeled and parameterized using the PSSM. We used KRAS, a well-characterized oncogene, as an example of how model-based analysis performs using consensus data to identify low allele frequency variants (see Fig. 1). There is a clear separation of background reads and reads positive for KRAS p.G12R (Fig. 1c). The results show that we were able to obtain statistical significance below 0.1% VAF for this variant (Fig. 1c). This oncogenic hotspot regularly shows 0 variant reads for negative samples, and for positive samples, at least ten reads supporting the G12R are detected. If P(R|M) < α, then variant is considered a call candidate. Figure 1d shows an example of the p values for calling the variant KRAS p.G12R, which has an α of 0.01. The points marked in red indicate samples that were called positive, while blue demarks samples called negative.
To demonstrate the dynamic range of SNV detection, DNA samples from multiple cell lines were mixed at various ratios to create two sample pools with a total of 188 variants, with allele frequencies ranging from 0.5 to 100% for each pool (see the supplementary methods for details). We identify 188 out of 188 expected variants, with allele frequencies within the expected range (Fig. 2a). To establish the LOD, we used reference cfDNA samples which contain 289 known clinically relevant SNVs with a range of allele frequencies from 0.1 to 5%. Figure 2b shows the comparison between observed and expected allele frequencies for the SNVs. Across 289 known variants, we identify 90 out of 106 variants (84.9%) with an allele frequency ≤ 0.2% and 183 out of 183 variants (100%) with an allele frequency > 0.2%.
Detection of single-nucleotide variants (SNVs) and assay reproducibility. a Comparison of observed and expected allele frequencies for 188 SNVs from two different sample pools with expected variant allele fractions (VAFs) from 0.5% to 100%. Pools were created by mixing 4 cell lines mixed at various ratios to create variants with a wide range of allele frequencies. b Comparison of observed and expected allele frequencies of multiple sample sets with expected VAFs ranging from 0.05% to 5%. The samples sets are made of Horizon cell-free DNA (cfDNA) reference samples with VAFs of 0.1, 1 and 5%, 2% Seracare cfDNA reference mixed with wild-type DNA to make dilutions of 0.1, 0.2, 0.5, 1 and 2%, and 10 cfDNA samples from healthy patients mixed at various ratios to produce sample pools with expected VAFs from 0.05% to 10%. Each sample set has 6, 23, and 75 unique variants, respectively. c Assessment of reproducibility and variability by assessment of observed allele frequencies for 6 variants from 29 replicate reference samples. Horizon cfDNA reference samples with VAFs of 0.1, 1 and 5% were sequenced 10, 10 and 9 times, respectively. For each of the expected VAFs, a boxplot shows the range of allele frequencies observed for each of the 6 variants
Commercially available reference samples and fragmented cell lines only mimic cfDNA and may mask assay problems due to variability in cfDNA size or complications from interfering substances. To show the test’s ability to detect variants from the intended sample matrix (cfDNA from plasma), whole blood was obtained from ten healthy donors with no history of cancer. gDNA and cfDNA were extracted for each patient sample. The gDNA was sent out to an external CLIA-CAP lab for whole-exome sequencing to establish each sample’s germline reference, and cfDNA was tested. We mixed cfDNA of the ten healthy samples at various ratios to create six sample pools with expected allele frequencies from 0.05 to 10%. Figure 2b shows the results comparing the observed allele frequency produced with the expected allele frequency for variants which were externally verified by an independent CLIA/CAP lab. We are able to correctly identify 23 out of 25 SNVs (92.0%) with allele frequency ≤ 0.2% and 52 out of 52 (100%) at > 0.2% from cfDNA extracted from plasma. The two variants not called were all observed at allele frequencies between 0.05 and 0.1%.
To demonstrate reproducibility, 36 libraries from cfDNA reference standards with 0, 0.1, 1 and 5% allele frequencies were sequenced seven, ten, ten and nine times, respectively. These libraries were run by different operators, on different days at various times over an extended time period. Each reference sample contained six ddPCR-verified SNVs in the panel’s 85-kb target regions. Figure 2c has boxplots showing the distribution of observed VAFs for each of the six variants for all positive samples. For negative reference samples (wild type), no false positives were observed after seven repeated library preparations and sequencing runs (data not shown). The error bars represent standard deviation, which is indicative of the variability in detection, and as can be observed, there is no significant difference, suggesting robust assay reproducibility.
SNV performance is summarized in Table 1. We are able to correctly identify 86.26% of variants with an allele frequency ≤ 0.2% and 100% of variants with an allele frequency > 0.2% with OncoLBx. A total of five false-positive variants were detected in two samples across all allele frequencies for over 1.8 million base pairs sequenced, providing a specificity of > 99.999%. We also demonstrated reproducibility and repeatability for SNV detection across allele frequencies ranging from 0.1 to 100%. In total, 536 out of 554 variants were correctly identified, giving 96.75% sensitivity, and five loci were called positive incorrectly, giving > 99.999% specificity and accuracy for identifying SNVs (see Sect. 2).
3.3 Indel Detection Performance
To establish LOD, we used cfDNA reference samples that contain 12 ddPCR-verified indels (seven insertions and five deletions) within OncoLBx’s target regions. We prepared samples by mixing reference sample with background wild type at 2, 1, 0.5, 0.2, 0.1 and 0%, with a total of 60 indel variants across all dilutions. Figure 3 shows the range of allele frequencies observed compared with their expected allele frequency for all the expected variants across the dilution series. In these samples, we were able to detect 56 out of 60 indels across all allele frequencies, and no false positives were detected in the 0% control sample.
Detection of insertions and deletions (indels). Comparison of the expected and observed variant allele fractions (VAFs) of 12 indels from cell-free DNA (cfDNA) references samples with VAFs ranging from 0% to 2%. Seracare cfDNA reference samples were mixed with wild-type cfDNA to create a dilution series with VAFs of 0, 0.1, 0.2, 0.5, 1, and 2%. Ins insertion, Del deletion
The dynamic range for detecting indels was tested using breast cancer cell lines from Coriell, with known BRCA1/2 indels combined at specific ratios into two different pools. Across both pools, there were a total of eight small indel variants (< 10 bp) with allele frequencies ranging from 0.5 to 20% and a 40-bp large deletion in BRCA1. Table 2 displays the known indel variants and compares their expected versus observed allele frequencies. We are able to accurately identify five out of five variants in BRCA1 and four out of four in BRCA2. The large BRCA1 40-base deletion (c.1175_1214del40) showed lower allele frequency than expected, but was still called correctly (Table 2). In addition, we also tested a cell line with a known 15-base EGFR exon 19 deletion, with expected variant frequencies ranging from 30 to 0.1%. We are able to call this deletion down to 0.3% with high confidence, and while detected at 0.1%, the variant did not meet our requirements for a confident call. Based on these results, we are able to call short indels (< 10 bp) with allele frequencies as low as 0.1% and large indels at least as low as 0.3%, with results indicating detection at lower allele frequencies is possible, but may have reduced sensitivity compared with short indels and SNVs.
Overall indel performance was assessed by aggregating the results for these two data sets and determining positive and negatives for both in Table 1. As shown in Table 1, variants with allele frequencies above 0.2% are identified with 100% sensitivity and specificity, whereas for VAFs ranging from 0.1 to 0.2%, the sensitivity and specificity are 83.33% and 99.999% respectively. These data demonstrate the ability to identify clinically relevant variants and variants with a LOD as low as 0.1%.
3.4 CNV Detection Performance
We validated CNV detection using OncoLBx by testing 33 reference samples and cell lines with known copy number amplifications (CNAs). Using HCC2218, with a known ERBB2 CNA, we created a dilution series with VAFs of 100%, 50%, 20%, 10%, 5% and 0%. Figure 4a shows the log ratio of counts between each of the HCC2218 dilution samples and a normal healthy sample. We identified amplifications in all exons of ERBB2, representing > 21 kb of chromosome 17, across all positive variant fractions (5–100%).
Detection of copy number variants (CNVs), gene fusions and microsatellite instability (MSI). a Dilution series of a cell line with a known ERBB2 copy number amplification visualized by comparing the log ratio of normalized coverage values between a sample and a healthy control. DNA from the breast cancer cell line HCC2218 was mixed with wild type to create samples with tumor fractions of 100, 50, 10, 5, and 0% (0% not shown). b Graph showing the EGFR amplification from the HCC827 cell line, plotting known copy number against observed counts (on a log scale). Copy numbers 14.6 and lower were done in triplicate. c A boxplot showing the reproducibility of detecting 4.5 copies across 11 control reference samples and healthy controls. d A dilution series of samples with a known fusion visualized with Integrated Genomics Viewer (IGV). DNA from the lung cancer cell line (H2228) with a known EML4–ALK fusion was spiked into wild type to create 6 samples with expected variant allele fractions (VAFs) of 0, 0.5, 1, 5, 10 and 100%. Aligned reads at the fusion breakpoint are shown for each fusion partner; ALK is shown at 100%, and the inset shows EML4’s breakpoint for each sample in the series. e DNA from the MSI-high colorectal cancer cell line HCT116 was mixed with wild-type DNA to create samples at various tumor fractions. The figure shows the number of reads at one marker with specific deletion size for samples with 10, 5, 2.5 and 0% tumor fractions. Peaks present in samples with positive tumor fractions were not detected in wild type
We validated the ability to detect a dynamic range of copy numbers using a reference sample (HCC827) with a verified 65-copy EGFR amplification spanning 17 kb of chromosome 7. Table 3 shows the copy number dilution series of EGFR and the number of replicates run for each copy number. All 11 samples had their estimated counts compared against expected, as plotted in Fig. 4b. We are able to detect EGFR amplifications in all four exons covered in the panel for each expected copy number representing the range of variant fractions from 100 to 5%. We are also able to identify the 2-copy (healthy) control samples and correctly call them as CNA negative.
Reproducibility was tested using the Horizon structural reference sample. This sample has been verified using ddPCR to contain 4.5 copies of MET with a whole gene amplification (~ 97 kb of chromosome 7). We tested 11 replicates of the sample, and we detected amplification of each exon in MET in all 11 replicates. In Fig. 4c, we show a boxplot of detected values for all reference sample replicates and compare it against another boxplot made of detected counts from healthy control samples (2 copies). These results demonstrate that we can reproducibly detect CNA of 4.5 copies and define the LOD as ≥ 4.5 copies.
3.5 Fusion Detection Performance
Fusions are identified by split read analysis covering all genes in the panel, with a focus on genes with known treatment guidelines. Single molecule split reads are used to estimate breakpoints and identify fusions (see Sect. 2). Since it is difficult to obtain reference samples with validated fusions, the validation is performed using all commercially available samples with four verified fusions. The ability to detect fusions was validated using cell lines and reference control samples with known fusions targeting RET-CCDC6, EML4-ALK, NCOA4-RET and TPR-ALK (see the supplementary methods in the ESM). The cell line containing the EML4-ALK fusion was tested at dilutions of 100, 10, 5, 1 and 0.5% to determine dynamic range. Figure 4d shows the visualization of the reads detected spanning ALK and EML4 at each dilution. Using OncoLBx we were able to correctly identify the fusion at all allele frequencies. One reference sample containing two fusions (NCOA4-RET and TPR-ALK) was tested at frequencies of 2, 1, 0.5, 0.2 and 0.1% to determine LOD. OncoLBx correctly identified both fusions in all samples; however, lower allele frequency samples (≤ 0.2%) had low coverage, reducing calling confidence for these samples. Overall, we were able to correctly identify 100% of fusion variants with high confidence at VAFs as low as 0.5% (Table 1), and these can be detected at lower VAFs, but sensitivity may be reduced.
3.6 MSI Detection Performance
MSI status is determined by analysis of length changes in five repeat regions (see Sect. 2). Samples with two or more unstable sites are deemed MSI-H. DNA from MSI-H cell lines was spiked into normal DNA and fragmented to mimic cfDNA and tested with OncoLBx. Figure 4e shows the number of reads showing deletions of a specific size, ranging from 1 to 30 bases, for HCT-116 spike-in samples at one repeat region. Deletions observable in all MSI-H samples were not detected in wild type. The LOD for MSI was tested with serial dilutions of HCT-15 and HCT-116 fragmented DNA into background DNA. MSI status was correctly determined with VAFs of 2.5% for HCT-116 and 2% for HCT-15, demonstrating the ability to detect MSI with tumor fractions as low as 2%. All seven samples were correctly identified as MSI-H for all tumor fractions in both cell lines. Table 1 summarizes the analytical results, showing 100% sensitivity and specificity for tumor fractions as low as 2%.
3.7 Clinical Validation
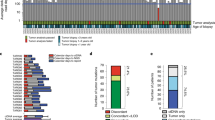
We assessed clinical performance using 36 clinical samples for which both blood and tissue formalin-fixed paraffin-embedded (FFPE) were available. These samples were sourced from patients diagnosed with stage II, III, or IV colorectal cancer (seven) and lung adenocarcinoma (29). For NGS-based tissue/liquid biopsy tests, the established methodology to evaluate diagnostic performance is a comparison of variants detected in blood with variants detected in tissue (FFPE) samples [19]. For clinical validation, the diagnostic performance was determined by designating variants detected in one sample type (tissue or blood) as the reference call set and then evaluating the performance of the variant identification using the other sample type. This was then repeated with the sample types reversed. When using the FFPE sample variants as the reference set, liquid biopsy achieved a clinical sensitivity of 80.22% (Table 4). In the opposite case, with cfDNA calls used as the reference call set, FFPE samples had a clinical sensitivity of 82% (Table 4). A total of 73 variants were identified across all 36 sample pairs (average of 2.02 variants per sample), including four samples with fusions (EML4–ALK) and eight samples with amplifications (EGFR and PDGFRA). The most commonly mutated genes were EGFR, APC, KRAS and TP53. Figure 5c visualizes the EGFR amplification from a paired sample with the variant detected in both tissue and blood. Figure 5d is a visualization of the breakpoint coverage of the EML4-ALK fusion detected in a blood and tissue sample pair. Five FFPE samples with enough genetic material for variant confirmation were sent for Sanger sequencing. Sanger confirmed the presence of all five targeted variants that were called in the FFPE samples.
Clinical validation. a Diagnosed cancer types and stage for 416 clinical samples. Breast, colon, and lung were the most common cancer types. Most of the samples were stage IV, and 77% of the samples stage III/IV. b The percentage of clinical samples with actionable variants detected, grouped by stage. c Comparison of an EGFR copy number variant identified in paired blood and formalin-fixed paraffin-embedded (FFPE) tissue. The log ratio of normalized coverage between each sample and a healthy control is shown for both sample types. d Comparison of EML4–ALK fusion identified in a paired blood and FFPE sample. The coverage of the breakpoint in both genes is shown, constructed to show the resulting fusion DNA sequence
In total, blood from 416 patients from a Taiwan cohort were tested with OncoLBx. Figure 5a shows the distribution of primary tumor types from the patient sample group, with the most common being colon, lung, breast, ovarian and pancreatic cancers. We identified somatic mutations (positive) in 79% of samples, with 87.34% of these samples having clinically relevant variants (variants with known oncological effects). Across all tumor types, TP53 was the most commonly mutated gene, followed by APC, PTEN, BRCA1/2 and PIK3CA. Over half of the samples (56%) were stage IV, 21% stage III, and 24% stage I/II (Fig. 5a). Actionable variants (variants with known targeted treatment options) were identified across all disease stages: 50% of stage I, 62% of stage II, 89% of stage III and 86% of stage IV patients (Fig. 5b). A total of 76 samples were tested for fusions, with nine out of 76 (12%) showing fusions, eight of which were EML4-ALK fusions, and 1 was a FGFR3-TACC3 fusion. In 55 clinical samples diagnosed with lung adenocarcinoma, EGFR variants were detected in 25 out of 55 samples (45%), RAS variants in five out of 55 samples (9%), and a BRAF variant in one out of 55 samples (2%). In addition, fusions were detected in three out of 55 samples and amplifications were detected in six out of 55 samples, which are consistent with previously published mutation frequencies [36,37,38,39,40]. The most common EGFR variants, in descending order, were p.L858R, ex19 deletion, p.T790M and ex20 insertions.
4 Discussion
NGS-based genomic profiling has been demonstrated to be effective in helping identify relevant targeted and immunotherapies to improve and manage cancer patient care [2, 4,5,6, 41, 42]. OncoLBx allows the assessment of all NCCN guideline-relevant treatments with known genomic variants, using peripheral blood from cancer patients with solid tumors. We demonstrated that OncoLBx paired with SMSEQ has high sensitivity and specificity across a large range of variant frequencies (VAF 0.1–100%) for SNVs, indels, CNAs, fusions, and MSI. The SMSEQ platform improves the signal-to-noise ratio for detecting genetic alterations at ultra-low allele frequencies in ctDNA, and corrects NGS artifacts and bias created through library preparation and sequencing. Consistently accurate detection of a range of variant types is aided by stringent QC steps included in the platform. The error correction process enables variant calling with high sensitivity, specificity, and accuracy.
CAP/AMP guidelines for validation of an NGS clinical assay include defining real and effective QC metrics, adequate statistical power to support claims and accurate determination of test performance [33]. Strict QC steps become important for high-sensitivity tests like liquid biopsy to ensure consistent and reliable results. We have developed stepwise QC metrics and stringent acceptance criteria to monitor and guide sample processing from sample preparation to variant calling (see Sect. 2 and supplementary Figure S1). The establishment of QC metrics/acceptance criteria allows us to generate consistent high-quality data, which is essential for identifying variants with low VAFs. For a high-sensitivity assay with large dynamic ranges, such as OncoLBx, more reference samples are required to demonstrate capability and robustness of calling low allele frequency. We used over 126 reference and cell-line samples and hundreds of clinical samples to validate OncoLBx. Without this rigorous characterization of a test, issues such as sampling bias could have a significant effect on test accuracy.
The challenge of identifying variants at low VAFs is different for each variant type. For SNVs, calling variants near the error rate of the Illumina platform was primarily solved through the use of SMSEQ and the development of algorithms trained to specifically identify SNVs (Fig. 1). Run-to-run variability was found to have no impact on variants with VAFs > 0.2%; however, results suggest that increased variability at lower allele frequencies (< 0.2%) may increase the chance of incorrect variant calling. Target capture bias did not appear to play a significant role in SNV detection, but may be a factor in performance of larger structural variants, such as fusions and long indels (see Sect. 3). This difference in performance between short and long indels in a capture-based assay is consistent with previous work showing that bias increases as the difference between sequencing read and capture sequence increases [40]. Fusion breakpoints also showed a lower observed allele frequency compared to expected. To confirm this hypothesis, we designed amplicons with the established breakpoint at various positions in the amplicon to compare capture performance based on breakpoint position (data not shown). From this testing, we confirmed that as we moved the breakpoint so that fewer bases covered the capture probe target region, performance (reads detected) dropped significantly. Increasing probe concentration or adjusting probe tiling strategies in low coverage regions may remediate the issue.
Our results showed that OncoLBx is highly concordant with tissue-based typing; however, when comparing paired blood and tissue samples, some sample pairs were discordant. We believe that variants detected in blood and not FFPE samples are mostly a result of tumor heterogeneity and clonal mutation in metastatic sites [21,22,23,24,25]. We also believe that variants found in FFPE samples, but not blood, are because not all tumors shed DNA at the same rate, if at all [26,27,28,29,30]. Our results show the assay’s ability to detect variants correlated with disease progression; cfDNA testing was less likely to detect variants in samples classified as stage II (60%) and more likely (> 85%) in stages III and IV. This correlates well with discordance between blood and tissue.
There are limitations to the OncoLBx assay. Most prominent is that not all tumors and stages will shed ctDNA at levels that are detectable. Additionally, the scope of this validation does not include copy number losses. While OncoLBx can detect copy number losses, analytically it is difficult to determine whether there is a loss of one or both copies. For therapy matching purposes, amplifications typically have more positive treatment indications. Another limitation is OncoLBx only targets known fusion sites, with NCCN-guideline treatments. There are known (less-common) fusion sites that are not covered by the panel. Finally, this validation study was limited by only having paired colon and lung cancer samples for comparison. Despite this, this study as a whole demonstrates that OncoLBx can detect five different variant types at low allele frequencies regardless of cancer type and meets guidelines for calling all five variant types for all cancers [33].
In addition to validation with peripheral blood, we have previously tested OncoLBx with other body fluids known to harbor ctDNA, such as pleural effusion and cerebral spinal fluid [43,44,45,46]. In many of these cases, we are able to identify variants with therapy options from the non-blood body fluids. In the case of cerebral spinal fluid, we were able to identify a targeted therapy option which had an optimal treatment outcome.
Accurate real-time, non-invasive tumor typing can help physicians to optimize and monitor the treatment of cancer patients [3, 13]. OncoLBx has been developed and validated as a liquid biopsy assay for identifying tumor variants at low allele frequencies from peripheral blood, with the ability to detect actionable variants from a wide range of solid tumor types and stages. It is an alternate testing platform available to doctors and patients to provide a non-invasive typing option. While not part of this validation, we believe that OncoLBx can be used to monitor tumor evolution in real-time, and disease progress may be trackable across multiple tests to monitor for treatment efficacy and disease progression.
References
Cummings CA, Peters E, Lacroix L, Andre F, Lackner MR. The role of next-generation sequencing in enabling personalized oncology therapy. Clin Transl Sci. 2016;9(6):283–92.
Lindeman NI, Cagle PT, Aisner DL, Arcila ME, Beasley MB, Bernicker E, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med. 2018;142:321–46.
Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–58.
Boyiadzis MM, Kirkwood JM, Marshall JL, Pritchard CC, Azad NS, Gulley JL. Significance and implications of FDA approval of pembrolizumab for biomarker-defined disease. J Immunother Cancer. 2018;6(1):1–7.
Bonneville R, Krook MA, Kautto EA, Miya J, Wing MR, Chen H-Z, et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol. 2017;1(1):1–15.
Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–13.
Horak P, Fröhling S, Glimm H. Integrating next-generation sequencing into clinical oncology: strategies, promises and pitfalls. ESMO Open. 2016;1(5):e000094.
Barlesi F, Mazieres J, Merlio J-P, Debieuvre D, Mosser J, Lena H, et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet. 2016;387(10026):1415–26.
Carter J, Illei P, Zheng G, Lin M-T, Tseng L-H, Dudley J, et al. Non-p.V600E BRAF mutations are common using a more sensitive and broad detection tool. Am J Clin Pathol. 2015;144(4):620–8.
Planchard D, Smit EF, Groen HJM, Mazieres J, Besse B, Helland Å, et al. Dabrafenib plus trametinib in patients with previously untreated BRAFV600E-mutant metastatic non-small-cell lung cancer: an open-label, phase 2 trial. Lancet Oncol. 2017;18(10):1307–16.
Murtaza M, Dawson S-J, Tsui DWY, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497(7447):108–12.
Sausen M, Phallen J, Adleff V, Jones S, Leary RJ, Barrett MT, et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun. 2015;6:7686.
Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–92.
Overman MJ, Modak J, Kopetz S, Murthy R, Yao JC, Hicks ME, et al. Use of research biopsies in clinical trials: are risks and benefits adequately discussed? J Clin Oncol. 2013;31(1):17–22.
Yates LR, Knappskog S, Wedge D, Tutt A, Lønning PE, Campbell PJ. Genomic evolution of breast cancer metastasis and article genomic evolution of breast cancer metastasis and relapse. 2017;32:169–84.
Hong MKH, Macintyre G, Wedge DC, Van Loo P, Patel K, Lunke S, et al. Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer. Nat Commun. 2015;6:6605.
Todenhöfer T, Struss WJ, Seiler R, Wyatt AW, Black PC. Liquid biopsy-analysis of circulating tumor DNA (ctDNA) in Bladder Cancer. Bl Cancer. 2018;4(1):19–29.
Swarup V, Rajeswari MR. Circulating (cell-free) nucleic acids—a promising, non-invasive tool for early detection of several human diseases. FEBS Lett. 2007;581(5):795–9.
Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10(8):472–84.
Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One. 2015;10(10):1–27.
Chan KCA, Jiang P, Zheng YWL, Liao GJW, Sun H, Wong J, et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem. 2013;59(1):211–24.
Chan KCA, Hung ECW, Woo JKS, Chan PKS, Leung S-F, Lai FPT, et al. Early detection of nasopharyngeal carcinoma by plasma Epstein-Barr virus DNA analysis in a surveillance program. Cancer. 2013;119(10):1838–44.
Cheng F, Su L, Qian C. Circulating tumor DNA: a promising biomarker in the liquid biopsy of cancer. Oncotarget. 2015;7(30).
Bettegowda C, Sausen M, Leary R, Kinde I, Agrawal N, Bartlett B, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24.
Pantel K, Alix-Panabières C. Real-time liquid biopsy in cancer patients: fact or fiction? Cancer Res. 2013;73:6384–8.
Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579–86.
Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017. https://doi.org/10.1126/scitranslmed.aan2415.
Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–90.
Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci USA. 2005;102(45):16368–73.
Newman AM, Bratman SV, To J, Wynne JF, Eclov NCW, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20(5):548–54.
Gregory MT, Bertout JA, Ericson NG, Taylor SD, Mukherjee R, Robins HS, et al. Targeted single molecule mutation detection with massively parallel sequencing. Nucleic Acids Res. 2016;44(3):1–11.
Park G, Park JK, Shin SH, Jeon HJ, Kim NKD, Kim YJ, et al. Characterization of background noise in capture-based targeted sequencing data. Genome Biol. 2017;18(1):1–13.
Jennings LJ, Arcila ME, Corless C, Kamel-Reid S, Lubin IM, Pfeifer J, et al. Guidelines for validation of next-generation sequencing–based oncology panels: a joint consensus recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn. 2017;19(3):341–65.
Hempelmann JA, Scroggins SM, Pritchard CC, Salipante SJ. MSI plus for integrated colorectal cancer molecular testing by next-generation sequencing. J Mol Diagn. 2015;17(6):705–14.
Suraweera N, Duval A, Reperant M, Vaury C, Furlan D, Leroy K, et al. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology. 2002;123(6):1804–11.
Paez JG. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science (N Y). 2004;304(5676):1497–500.
Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci. 2004;101(36):13306–11.
Paik PK, Arcila ME, Fara M, Sima CS, Miller VA, Kris MG, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol. 2011;29(15):2046–51.
Pratilas CA, Hanrahan AJ, Halilovic E, Persaud Y, Soh J, Chitale D, et al. Genetic predictors of MEK dependence in non-small cell lung cancer. Cancer Res. 2008;68(22):9375–83.
Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000.
Mohamed A, Krajewski K, Cakar B, Ma CX. Targeted therapy for breast cancer. Am J Pathol. 2013;183(4):1096–112.
Masoud V, Pagès G. Targeted therapies in breast cancer: new challenges to fight against resistance. World J Clin Oncol. 2017;8(2):120.
Huang WT, Lu NM, Hsu WY, Chang SE, Atkins A, Mei R, et al. CSF-ctDNA SMSEQ analysis to tailor the treatment of a patient with brain metastases: a case report. Case Rep Oncol. 2018;11(1):68–74.
Li Y, Pan W, Connolly ID, Reddy S, Nagpal S, Quake S, et al. Tumor DNA in cerebral spinal fluid reflects clinical course in a patient with melanoma leptomeningeal brain metastases. J Neurooncol. 2016;128(1):93–100.
Husain H, Nykin D, Bui N, Quan D, Gomez G, Woodward B, et al. Cell-free DNA from ascites and pleural effusions: molecular insights into genomic aberrations and disease biology. Mol Cancer Ther. 2017;16(5):948–55.
Peng M, Chen C, Hulbert A, Brock MV, Yu F. Non-blood circulating tumor DNA detection in cancer. Oncotarget. 2017;8(40):69162–73.
Acknowledgements
We would like to acknowledge Medical Technologists of CellMax Life Taiwan Clinical Lab for processing the clinical samples. Especially, thanks to Shih-En Chang, Jr-Ming Lai and Twinkal Marfatia for coordinating clinical sample collection and maintaining the clinical database. This work would not have been possible without clinical samples provided by doctors from various medical facilities and hospitals in Taiwan; we would like to thank Dr. Chia-Hsun Hsieh (Chang Gung Memorial Hospital), Dr. Po-Jen Liu (Elite Clinic), Dr. Jui-Kun Hsieh (Evergreen Health), Dr. Yu-Lin Lin (National Taiwan University Hospital), Dr. Hung-Chih Lai (Shin Kong Hospital), and Dr. Chung-Ken Wu (Taichung). We would also like to thank Cardinal Tien Hospital, China Medical University Hospital, Chung Shan Medical University Hospital, Kaohsiung Veterans General Hospital, Taipei Veterans General Hospital, Taichung Veterans General Hospital, Tung’s Taichung Metro Harbor Hospital, and Core (India).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
AA., P.G., J.L., M.J., A.V. and R.M. are employed at CellMax Life, which provided funds for this research. B.M.Z. and W.T. report no known conflicts of interest.
Funding
The research was carried out at CellMax Life, which provided the funds.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
SMSEQ Assay Workflow and Select Quality Control Metrics. The figure is a diagrammatic representation of the workflow of the SMSEQ process consisting of five major steps: extraction, library preparation, target enrichment, sequencing and data analysis. There are multiple quality control steps and checkpoints throughout the workflow ensuring accurate detection of variants by targeted sequencing of libraries prepared from cell-free DNA extracted from plasma of patients. QC1: verifying cfDNA concentration, QC2: verifying cfDNA size and cfDNA/gDNA ratio, QC3: verifying library yield (400-800 ng), QC4: verifying library size-distribution (200-850 bp), QC5: verifying enriched libraries yield by QPCR, QC6: verifying cluster density, QC7: monitoring average coverage (> 10,000X) and QC8: determining off-target rate (< 30%). The data analysis step further includes multiple quality control parameters such as single molecule quality assessment, quality filtering etc. to obtain accurate data (TIFF 7273 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Atkins, A., Gupta, P., Zhang, B.M. et al. Detection of Circulating Tumor DNA with a Single-Molecule Sequencing Analysis Validated for Targeted and Immunotherapy Selection. Mol Diagn Ther 23, 521–535 (2019). https://doi.org/10.1007/s40291-019-00406-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-019-00406-0