Abstract
Background
Traditional methods for rejection control in transplanted patients are considered invasive, risky, and prone to sampling errors. Using molecular biomarkers as an alternative protocol to biopsies, for monitoring rejection may help to mitigate some of these problems, increasing the survival rates and well-being of patients. Recent advances in omics technologies provide an opportunity for screening new molecular biomarkers to identify those with clinical utility.
Objective
This systematic literature review (SLR) aimed to summarize existing evidence derived from large-scale expression profiling regarding differentially expressed mRNA and miRNA in graft rejection, highlighting potential molecular biomarkers in transplantation.
Methods
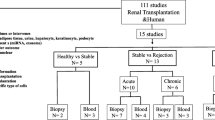
The study was conducted following PRISMA methodology and the BiSLR guide for performing SLR in bioinformatics. PubMed, ScienceDirect, and EMBASE were searched for publications from January 2001 to January 2018, and studies (i) aiming at the identification of transplant rejection biomarkers, (ii) including human subjects, and (iii) applying methodologies for differential expression analysis from large-scale expression profiling were considered eligible. Differential expression patterns reported for genes and miRNAs in rejection were summarized from both cross-organ and organ-specific perspectives, and pathways enrichment analysis was performed for candidate biomarkers to interrogate their functional role in transplant rejection.
Results
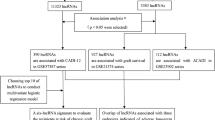
A total of 821 references were collected, resulting in 604 studies after removal of duplicates. After application of inclusion and exclusion criteria, 33 studies were included in our analysis. Among the 1517 genes and 174 miRNAs identifed, CXCL9, CXCL10, STAT1, hsa-miR-142-3p, and hsa-miR-155 appeared to be particularly promising as biomarkers in transplantation, with an increased expression associated with transplant rejection in multiple organs. In addition, hsa-miR-28-5p was consistently decreased in samples taken from rejected organs.
Conclusion
Despite the need for further research to fill existing knowledge gaps, transcriptomic technologies have a relevant role in the discovery of accurate biomarkers for transplant rejection diagnostics. Studies have reported consistent evidence of differential expression associated with transplant rejection, although issues such as experimental heterogeneity hinder a more systematic characterization of observed molecular changes. Special attention has been giving to large-scale mRNA expression profiling in rejection, whereas there is still room for improvements in the characterization of miRnome in this condition.
PROSPERO Registration Number
CRD42018083321.





Similar content being viewed by others
References
Khatri P, Roedder S, Kimura N, et al. A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation. J Exp Med. 2013;210:2205–21.
Rana A, Ackah RL, Webb GJ, et al. No gains in long-term survival after liver transplantation over the past three decades. Ann Surg. 2019;269:20–7.
Wang A, Sarwal MM. Computational models for transplant biomarker discovery. Front Immunol. 2015;6:458.
Wilhelm MJ. Long-term outcome following heart transplantation: current perspective. J Thorac Dis. 2015;7:549–51.
Ettenger R, Albrecht R, Alloway R. Meeting report: FDA public meeting on patient-focused drug development and medication adherence in solid organ transplant patients. Am J Transplant. 2018;18(3):564–73.
Baron D, Ramstein G, Chesneau M, et al. A common gene signature across multiple studies relate biomarkers and functional regulation in tolerance to renal allograft. Kidney Int. 2015;87:984–95.
Loftheim H, Midtvedt K, Hartmann A, et al. Urinary proteomic shotgun approach for identification of potential acute rejection biomarkers in renal transplant recipients. Transplant Res. 2012;1:9.
Einecke G, Reeve J, Sis B, et al. A molecular classifier for predicting future graft loss in late kidney transplant biopsies. J Clin Invest. 2010;120:1862–72.
Burke HB. Predicting clinical outcomes using molecular biomarkers. Biomark Cancer. 2016;8:89–99.
Mas VR, Dumur CI, Scian MJ, et al. MicroRNAs as biomarkers in solid organ transplantation. Am J Transplant. 2013;13:11–9.
Sirota M, Sarwal MM. Transplantomics: toward precision medicine in transplantation research. Transplantation. 2017;101:1777–82.
Naesens M, Sarwal MM. Molecular diagnostics in transplantation. Nat Rev Nephrol. 2010;6:614–28.
Ritchie MD, Holzinger ER, Li R, et al. Methods of integrating data to uncover genotype-phenotype interactions. Nat Rev Genet. 2015;16:85–97.
Mariano DCB, Leite C, Santos LHS, et al. A guide to performing systematicQuery literature reviews in bioinformatics. 2017. Technical report—RT.DCC.002/2017. arXiv [q-bio.QM]. 2017. https://arxiv.org/abs/1707.05813. Accessed 25 Mar 2019.
Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535.
Wain HM, Bruford EA, Lovering RC, et al. Guidelines for human gene nomenclature. Genomics. 2002;79:464–70.
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–61.
Downs SH, Black N. The feasibility of creating a checklist for the assessment of the methodological quality both of randomised and non-randomised studies of health care interventions. J Epidemiol Community Health. 1998;52:377–84.
Flechner SM, Kurian SM, Head SR, et al. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant. 2004;4:1475–89.
Chen R, Sigdel TK, Li L, et al. Differentially expressed RNA from public microarray data identifies serum protein biomarkers for cross-organ transplant rejection and other conditions. PLoS Comput Biol. 2010;6(9):e1000940. https://doi.org/10.1371/journal.pcbi.1000940.
Gregson AL, Hoji A, Injean P, et al. Altered exosomal RNA profiles in bronchoalveolar lavage from lung transplants with acute rejection. Am J Respir Crit Care Med. 2015;192:1490–503.
Gimino VJ, Lande JD, Berryman TR, et al. Gene expression profiling of bronchoalveolar lavage cells in acute lung rejection. Am J Respir Crit Care Med. 2003;168:1237–42.
Lu BS, Yu AD, Zhu X, et al. Sequential gene expression profiling in lung transplant recipients with chronic rejection. Chest. 2006;130:847–54.
Chen Y, Zhang H, Xiao X, et al. Peripheral blood transcriptome sequencing reveals rejection-relevant genes in long-term heart transplantation. Int J Cardiol. 2013;168:2726–33.
Lin D, Hollander Z, Ng RT, et al. Whole blood genomic biomarkers of acute cardiac allograft rejection. J Heart Lung Transplant. 2009;28:927–35.
Alakulppi N, Seikku P, Jaatinen T, et al. Feasibility of diagnosing subclinical renal allograft rejection in children by whole blood gene expression analysis. Transplantation. 2008;86:1222–8.
Hollander Z, Lin D, Chen V, et al. Whole blood biomarkers of acute cardiac allograft rejection: double-crossing the biopsy. Transplantation. 2010;90:1388–93.
Karason K, Jernås M, Hägg DA, Svensson P-A. Evaluation of CXCL9 and CXCL10 as circulating biomarkers of human cardiac allograft rejection. BMC Cardiovasc Disord. 2006;6:29.
Vitalone MJ, Sigdel TK, Salomonis N, et al. Transcriptional perturbations in graft rejection. Transplantation. 2015;99:1882–93.
Wilflingseder J, Regele H, Perco P, et al. miRNA profiling discriminates types of rejection and injury in human renal allografts. Transplantation. 2013;95:835–41.
Scherer A, Krause A, Walker JR, et al. Early prognosis of the development of renal chronic allograft rejection by gene expression profiling of human protocol biopsies. Transplantation. 2003;75:1323–30.
Rascio F, Pontrelli P, Accetturo M, et al. A type I interferon signature characterizes chronic antibody-mediated rejection in kidney transplantation. J Pathol. 2015;237:72–84.
Horwitz PA, Tsai EJ, Putt ME, et al. Detection of cardiac allograft rejection and response to immunosuppressive therapy with peripheral blood gene expression. Circulation. 2004;110:3815–21.
Loupy A, Duong Van Huyen JP, Hidalgo L, et al. Gene expression profiling for the identification and classification of antibody-mediated heart rejection. Circulation. 2017;135:917–35.
Shen Z, Gong W. Identification of candidate biomarkers in peripheral blood for cardiac allograft rejection based on bioinformatics analysis. Ann Transplant. 2015;20:312–9.
Venner JM, Hidalgo LG, Famulski KS, et al. The molecular landscape of antibody-mediated kidney transplant rejection: evidence for NK involvement through CD16a Fc receptors. Am J Transplant. 2015;15:1336–48.
Talayero P, Alonso-Guirado L, Padilla G, et al. 5-Gene differential expression predicts stability of human intestinal allografts. Exp Mol Pathol. 2017;103:163–71.
Chen W, Peng W, Huang J, et al. Microarray analysis of long non-coding RNA expression in human acute rejection biopsy samples following renal transplantation. Mol Med Rep. 2014;10:2210–6.
Günther OP, Balshaw RF, Scherer A, et al. Functional genomic analysis of peripheral blood during early acute renal allograft rejection. Transplantation. 2009;88:942–51.
Sotolongo B, Asaoka T, Island E, et al. Gene expression profiling of microRNAs in small-bowel transplantation paraffin-embedded mucosal biopsy tissue. Transplant Proc. 2010;42:62–5.
Bodez D, Hocini H, Tchitchek N, et al. Myocardial gene expression profiling to predict and identify cardiac allograft acute cellular rejection: the GET-Study. PLoS One. 2016;11:e0167213.
Shannon CP, Hollander Z, Wilson-McManus J, et al. White blood cell differentials enrich whole blood expression data in the context of acute cardiac allograft rejection. Bioinform Biol Insights. 2012;6:49–61.
Anglicheau D, Sharma VK, Ding R, et al. MicroRNA expression profiles predictive of human renal allograft status. Proc Natl Acad Sci USA. 2009;106:5330–5.
Matz M, Fabritius K, Lorkowski C, et al. Identification of T cell–mediated vascular rejection after kidney transplantation by the combined measurement of 5 specific microRNAs in blood. Transplantation. 2016;100:898–907.
Asaoka T, Sotolongo B, Island ER, et al. MicroRNA signature of intestinal acute cellular rejection in formalin-fixed paraffin-embedded mucosal biopsies. Am J Transplant. 2012;12:458–68.
Liu X, Dong C, Jiang Z, et al. MicroRNA-10b downregulation mediates acute rejection of renal allografts by derepressing BCL2L11. Exp Cell Res. 2015;333:155–63.
Halloran PF, Famulski KS, Reeve J. Molecular assessment of disease states in kidney transplant biopsy samples. Nat Rev Nephrol. 2016;12:534–48.
Wu Z, Huang X, Han X, et al. The chemokine CXCL9 expression is associated with better prognosis for colorectal carcinoma patients. Biomed Pharmacother. 2016;78:8–13.
Lo DJ, Weaver TA, Kleiner DE, et al. Chemokines and their receptors in human renal allotransplantation. Transplantation. 2011;91:70–7.
Kumar S, Mohapatra N, Borle DP, et al. Non invasive diagnosis of acute cellular rejection after liver transplantation—current opinion. Transpl Immunol. 2018;47:1–9.
Chmielewski S, Piaszyk-Borychowska A, Wesoly J, Bluyssen HAR. STAT1 and IRF8 in vascular inflammation and cardiovascular disease: diagnostic and therapeutic potential. Int Rev Immunol. 2016;35:434–54.
Alegre M-L, Lakkis FG, Morelli AE. Antigen presentation in transplantation. Trends Immunol. 2016;37:831–43.
Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Front Immunol. 2014;5:461. https://doi.org/10.3389/fimmu.2014.00461.
Freue GVC, Cohen Freue GV, Sasaki M, et al. Proteomic signatures in plasma during early acute renal allograft rejection. Mol Cell Proteomics. 2010;9:1954–67.
Sukma Dewi I, Hollander Z, Lam KK, et al. Association of serum MiR-142-3p and MiR-101-3p levels with acute cellular rejection after heart transplantation. PLoS One. 2017;12:e0170842.
Seddiki N, Brezar V, Ruffin N, et al. Role of miR-155 in the regulation of lymphocyte immune function and disease. Immunology. 2014;142:32–8.
Duong Van Huyen J-P, Tible M, Gay A, et al. MicroRNAs as non-invasive biomarkers of heart transplant rejection. Eur Heart J. 2014;35:3194–202.
Danger R, Pallier A, Giral M, et al. Upregulation of miR-142-3p in peripheral blood mononuclear cells of operationally tolerant patients with a renal transplant. J Am Soc Nephrol. 2012;23:597–606.
Domenico TD, Joelsons G, Montenegro RM, Manfro RC. Upregulation of microRNA 142-3p in the peripheral blood and urinary cells of kidney transplant recipients with post-transplant graft dysfunction. Braz J Med Biol Res. 2017;50:e5533.
Anglicheau D, Naesens M, Essig M, et al. Establishing biomarkers in transplant medicine: a critical review of current approaches. Transplantation. 2016;100:2024–38.
Heidt S, San Segundo D, Shankar S, et al. Peripheral blood sampling for the detection of allograft rejection: biomarker identification and validation. Transplantation. 2011;92:1–9.
Paczesny S. Biomarkers for posttransplantation outcomes. Blood. 2018;131:2193–204.
Khush K, Zarafshar S. Molecular diagnostic testing in cardiac transplantation. Curr Cardiol Rep. 2017;19:118.
Nissaisorakarn V, Lee JR, Lubetzky M, Suthanthiran M. Urine biomarkers informative of human kidney allograft rejection and tolerance. Hum Immunol. 2018;79:343–55.
Asaoka T, Island ER, Tryphonopoulos P, et al. Characteristic immune, apoptosis and inflammatory gene profiles associated with intestinal acute cellular rejection in formalin-fixed paraffin-embedded mucosal biopsies. Transpl Int. 2011;24:697–707.
Deng MC, Eisen HJ, Mehra MR, et al. Noninvasive discrimination of rejection in cardiac allograft recipients using gene expression profiling. Am J Transplant. 2006;6:150–60.
Huang M-C, Tullo AB, Hillarby MC. Increased Rac2 mRNA expression in peripheral blood during human corneal graft rejection. Eye. 2009;23:461–9.
Acknowledgments
SVP and RHB acknowledge scholarships from CNPq (Brazilian National Council for Scientific and Technological Development).
Author information
Authors and Affiliations
Contributions
SVP and MR-M conceived and designed the study; SVP collected data; SVP, GHP, RHB, RC, and MR-M analyzed data; SVP and MR-M wrote the manuscript; GHP and RC critically reviewed the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Funding
No sources of funding were received for this systematic review.
Conflict of interest
SVP, GHP, RHB, RC, and MR-M declare that they have no conflicts of interest relevant to this systematic review.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Paladini, S.V., Pinto, G.H., Bueno, R.H. et al. Identification of Candidate Biomarkers for Transplant Rejection from Transcriptome Data: A Systematic Review. Mol Diagn Ther 23, 439–458 (2019). https://doi.org/10.1007/s40291-019-00397-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-019-00397-y