Abstract
Introduction
Intellectual disability (ID) is often sporadic, and its complex etiology makes clinical diagnosis extremely difficult.
Objective
The aims of this study were to detect copy number variations (CNVs) in patients with ID and to analyze the correlation between pathogenic CNVs and clinical phenotype.
Methods
After cases of ID caused by metabolic dysfunction or environmental factors were excluded, 64 patients with moderate to severe ID were enrolled. Karyotype and single nucleotide polymorphism (SNP) array analyses were performed for all patients. The relationship between CNVs and phenotype was identified with genotype–phenotype comparisons and by searching CNV databases.
Results
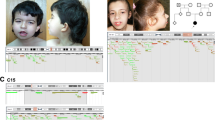
Karyotype analysis showed four patients with chromosomal aneuploidy and seven with chromosomal structural abnormality. After excluding the four cases with chromosomal aneuploidy, the remaining 60 cases were analyzed using SNP array. The results revealed 87 CNVs in 45 cases, including 16 pathogenic CNVs in 12 individuals, with a diagnostic yield of 20.0% (12/60). We found large deletions at 16q22.2q23.1 and 3q24q25.32 in two patients, respectively, in whom specific syndromes had not been defined. Our array analysis showed one case carried a 210 kb deletion at 1p21.2p21.3, which included only one coding gene LPPR4, which might be a candidate gene for ID phenotype.
Conclusions
Use of the genome-wide array method can improve the detection rate of CNVs, reveal chromosomal abnormalities that have not been well-characterized by cytology, and provide a new way to locate genes for patients with the ID phenotype. Interpretation of CNVs remains a major challenge. Sharing of CNVs and phenotype information from different laboratories in public databases is important.



Similar content being viewed by others
Change history
21 November 2018
An Online First version of this article was made available online.
21 November 2018
An Online First version of this article was made available online.
References
Moeschler JB, Shevell M. Comprehensive evaluation of the child with intellectual disability or global developmental delays. Pediatrics. 2014;134(3):e903–18. https://doi.org/10.1542/peds.2014-1839.
Honeycutt ACH, Homsi al G, Grosse S, Schendel D, Dunlap L. Economic costs associated with mental retardation, cerebral palsy, hearing loss, and vision impairment: United States, 2003. MMWR Morbidity Mortality Wkly Rep. 2004;53(3):57–9.
Vissers LE, de Vries BB, Osoegawa K, Janssen IM, Feuth T, Choy CO, et al. Array-based comparative genomic hybridization for the genomewide detection of submicroscopic chromosomal abnormalities. Am J Hum Genet. 2003;73(6):1261–70. https://doi.org/10.1086/379977.
de Vries BB, Pfundt R, Leisink M, Koolen DA, Vissers LE, Janssen IM, et al. Diagnostic genome profiling in mental retardation. Am J Hum Genet. 2005;77(4):606–16. https://doi.org/10.1086/491719.
Menten B, Maas N, Thienpont B, Buysse K, Vandesompele J, Melotte C, et al. Emerging patterns of cryptic chromosomal imbalance in patients with idiopathic mental retardation and multiple congenital anomalies: a new series of 140 patients and review of published reports. J Med Genet. 2006;43(8):625–33. https://doi.org/10.1136/jmg.2005.039453.
Fan YS, Jayakar P, Zhu H, Barbouth D, Sacharow S, Morales A, et al. Detection of pathogenic gene copy number variations in patients with mental retardation by genomewide oligonucleotide array comparative genomic hybridization. Hum Mutat. 2007;28(11):1124–32. https://doi.org/10.1002/humu.20581.
Moeschler JB. Genetic evaluation of intellectual disabilities. Semin Pediatr Neurol. 2008;15(1):2–9. https://doi.org/10.1016/j.spen.2008.01.002.
Manning M, Hudgins L, Professional P, Guidelines C. Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet Med Off J Am Coll Med Genet. 2010;12(11):742–5. https://doi.org/10.1097/GIM.0b013e3181f8baad.
Coe BP, Witherspoon K, Rosenfeld JA, van Bon BW, Vulto-van Silfhout AT, Bosco P, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet. 2014;46(10):1063–71. https://doi.org/10.1038/ng.3092.
Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86(5):749–64. https://doi.org/10.1016/j.ajhg.2010.04.006.
Gijsbers AC, Lew JY, Bosch CA, Schuurs-Hoeijmakers JH, van Haeringen A, den Hollander NS, et al. A new diagnostic workflow for patients with mental retardation and/or multiple congenital abnormalities: test arrays first. Eur J Hum Genet EJHG. 2009;17(11):1394–402. https://doi.org/10.1038/ejhg.2009.74.
Pradeep V, Mohnish S. A clinical approach to developmental delay and intellectual disability. Clin Med (Lond). 2017;17(6):558–61. https://doi.org/10.7861/clinmedicine.17-6-558.
Michelson DJ, Shevell MI, Sherr EH, Moeschler JB, Gropman AL, Ashwal S. Evidence report: Genetic and metabolic testing on children with global developmental delay: report of the Quality Standards Subcommittee of the American Academy of Neurology and the Practice Committee of the Child Neurology Society. Neurology. 2011;77(17):1629–35. https://doi.org/10.1212/WNL.0b013e3182345896.
Vissers LE, Stankiewicz P. Microdeletion and microduplication syndromes. Methods Mol Biol (Clifton, NJ). 2012;838:29–75. https://doi.org/10.1007/978-1-61779-507-7_2.
Caspersson T, Farber S, Foley GE, Kudynowski J, Modest EJ, Simonsson E, et al. Chemical differentiation along metaphase chromosomes. Exp Cell Res. 1968;49(1):219–22.
Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST. Working Group of the American College of Medical Genetics Laboratory Quality Assurance C. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genetics in medicine: official journal of the American College of Medical. Genetics. 2011;13(7):680–5. https://doi.org/10.1097/GIM.0b013e3182217a3a.
Brauer AU, Savaskan NE, Kuhn H, Prehn S, Ninnemann O, Nitsch R. A new phospholipid phosphatase, PRG-1, is involved in axon growth and regenerative sprouting. Nat Neurosci. 2003;6(6):572–8. https://doi.org/10.1038/nn1052.
Trimbuch T, Beed P, Vogt J, Schuchmann S, Maier N, Kintscher M, et al. Synaptic PRG-1 modulates excitatory transmission via lipid phosphate-mediated signaling. Cell. 2009;138(6):1222–35. https://doi.org/10.1016/j.cell.2009.06.050.
van Karnebeek CD, Jansweijer MC, Leenders AG, Offringa M, Hennekam RC. Diagnostic investigations in individuals with mental retardation: a systematic literature review of their usefulness. Eur J Hum Genet EJHG. 2005;13(1):6–25. https://doi.org/10.1038/sj.ejhg.5201279.
Schaefer GB, Bodensteiner JB. Evaluation of the child with idiopathic mental retardation. Pediatr Clin N Am. 1992;39(4):929–43.
Peters GB, Pertile MD. Chromosome microarrays in diagnostic testing: interpreting the genomic data. Methods Mol Biol (Clifton, NJ). 2014;1168:117–55. https://doi.org/10.1007/978-1-4939-0847-9_8.
Chen X, Li H, Mao Y, Xu X, Lv J, Zhou L, et al. Subtelomeric multiplex ligation-dependent probe amplification as a supplement for rapid prenatal detection of fetal chromosomal aberrations. Mol Cytogenet. 2014;7(1):96. https://doi.org/10.1186/s13039-014-0096-1.
De Gregori M, Ciccone R, Magini P, Pramparo T, Gimelli S, Messa J, et al. Cryptic deletions are a common finding in “balanced” reciprocal and complex chromosome rearrangements: a study of 59 patients. J Med Genet. 2007;44(12):750–62. https://doi.org/10.1136/jmg.2007.052787.
Zhu H, Hu Y, Zhu R, Yang Y, Zhu X, Wang W. A boy with partial trisomy of chromosome 3q24-q28 from paternal balanced insertion and multiple congenital anomalies. Am J Med Genet Part A. 2013;161A:327–30. https://doi.org/10.1002/ajmg.a.35637.
de Volo CP, Alfonsi M, Gatta V, Novelli A, Bernardini L, Fantasia D, Antonucci I, et al. 16q22.1 microdeletion detected by array-CGH in a family with mental retardation and lobular breast cancer. Gene. 2012;498(2):328–31. https://doi.org/10.1016/j.gene.2012.01.028.
Wright CF, Fitzgerald TW, Jones WD, Clayton S, McRae JF, van Kogelenberg M, et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015;385(9975):1305–14. https://doi.org/10.1016/S0140-6736(14)61705-0.
Acknowledgements
The authors thank the Genetic Counselling Clinic of Wenzhou Central Hospital for collecting patients’ phenotype information and Prof. Taosheng Huang from Cincinnati Children’s Hospital for revising this manuscript.
Author information
Authors and Affiliations
Contributions
ST initiated the study, coordinated clinical analyses of patients, and revised the manuscript. XC performed SNP array analyses and Q-PCR, and wrote the manuscript. HL and CC contributed to data collection and processing. XX helped write parts of the manuscript. LZ and YX performed cytogenetic analyses and cytogenetic diagnoses. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Xiangnan Chen, Huanzheng Li, Chong Chen, Lili Zhou, Xueqin Xu, Yanbao Xiang and Shaohua Tang have no conflicts of interest.
Funding
This study was funded by Wenzhou public technology and medical projects (Y20140655, Y20170207), Wenzhou technology bureau projects (ZD201302).
Informed Consent
Parents or guardians of the probands signed their written informed consent, which were approved by Dingli Clinical School Ethics Committee of Wenzhou Medical University.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, X., Li, H., Chen, C. et al. Genome-Wide Array Analysis Reveals Novel Genomic Regions and Candidate Gene for Intellectual Disability. Mol Diagn Ther 22, 749–757 (2018). https://doi.org/10.1007/s40291-018-0358-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-018-0358-4