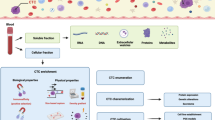
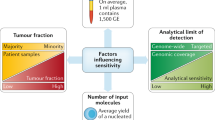
Abstract
There is a profound need in oncology to detect cancer earlier, guide individualized therapies, and better monitor progress during treatment. Currently, some of this information can be achieved through solid tissue biopsy and imaging. However, these techniques are limited because of the invasiveness of the procedure and the size of the tumor. A liquid biopsy can overcome these barriers as its non-invasive nature allows samples to be collected over time. Liquid biopsies may also allow earlier detection than traditional imaging. Liquid biopsies include the analysis of circulating tumor cells (CTCs), cell-free nucleic acid (cfNA), or extracellular vesicles obtained from a variety of biofluids, such as peripheral blood. In this review, we discuss different liquid biopsy types and how they fit into the current regulatory landscape.

Similar content being viewed by others
References
Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10(8):472–84.
Swanton C. Intratumor heterogeneity: evolution through space and time. Cancer Res. 2012;72(19):4875–82.
Gerlinger M, Rowan AJ, Horswell S, Larkin J, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–92.
Ashworth TR. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust Med J. 1896;14:146–7.
Mandel P, Metais P. C R Seances Soc Biol Fil. 1948;142(3–4):241–3.
Raj DA, et al. A multiplex quantitative proteomics strategy for protein biomarker studies in urinary exosomes. Kidney Int. 2012;81(12):1263–72.
Wiggins RC, et al. Procoagulant activity in normal human urine associated with subcellular particles. Kidney Int. 1986;29(2):591–7.
Asea A, et al. Heat shock protein-containing exosomes in mid-trimester amniotic fluids. J Reprod Immunol. 2008;79(1):12–7.
Witwer KW, Buzás EI, Bemis LT, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2. doi:10.3402/jev.v2i0.20360.
Caby MP, et al. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17(7):879–87.
Street JM, et al. Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J Transl Med. 2012;10:5.
Douillard JY, et al. First-line gefitinib in Caucasian EGFR mutation-positive NSCLC patients: a phase-IV, open-label, single-arm study. Br J Cancer. 2014;110(1):55–62.
Cristofanilli M, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351(8):781–91.
Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3(6):453–8.
Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 2004;21(2):137–48.
Nalejska E, Maczynska E, Lewandowska MA. Prognostic and predictive biomarkers: tools in personalized oncology. Mol Diagn Ther. 2014;18(3):273–84.
Hayes DF, et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res. 2006;12(14 Pt 1):4218–24.
Danila DC, et al. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13(23):7053–8.
Cohen SJ, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(19):3213–21.
Millner LM, Linder MW, Valdes R Jr. Circulating tumor cells: a review of present methods and the need to identify heterogeneous phenotypes. Ann Clin Lab Sci. 2013;43(3):295–304.
Gorges TM, et al. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer. 2012;12:178.
Vona G, et al. Isolation by size of epithelial tumor cells: a new method for the immunomorphological and molecular characterization of circulating tumor cells. Am J Pathol. 2000;156(1):57–63.
Ferreira MM, Ramani VC, Jeffrey SS. Circulating tumor cell technologies. Mol Oncol. 2016;10(3):374–94.
Fizazi K, et al. High detection rate of circulating tumor cells in blood of patients with prostate cancer using telomerase activity. Ann Oncol. 2007;18(3):518–21.
Stahlberg A, Bengtsson M. Single-cell gene expression profiling using reverse transcription quantitative real-time PCR. Methods. 2010;50(4):282–8.
Hodgkinson CL, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med. 2014;20(8):897–903.
Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–8.
Riethdorf S, et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the Cell Search system. Clin Cancer Res. 2007;13(3):920–8.
Schneck H, et al. EpCAM-independent enrichment of circulating tumor cells in metastatic breast cancer. PLoS One. 2015;10(12):e0144535.
Karabacak NM, et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat Protoc. 2014;9(3):694–710.
Maimonis P, et al. Poster no. 4898. American Association of Cancer Research 102nd Annual Meeting, Orlando, Florida, April 2–6 2011.
Gabriel MT, et al. Circulating tumor cells: a review of non-EpCAM-based approaches for cell enrichment and isolation. Clin Chem. 2016;62(4):571–81.
Vasioukhin V, et al. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br J Haematol. 1994;86(4):774–9.
Sorenson GD, et al. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomark Prev. 1994;3(1):67–71.
Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer: a survey. Biochim Biophys Acta. 2007;1775(1):181–232.
Medicine Kaiser J. Keeping tabs on tumor DNA. Science. 2010;327(5969):1074.
Higgins MJ, et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin Cancer Res. 2012;18(12):3462–9.
Castells A, et al. K-ras mutations in DNA extracted from the plasma of patients with pancreatic carcinoma: diagnostic utility and prognostic significance. J Clin Oncol. 1999;17(2):578–84.
Shinozaki M, et al. Utility of circulating B-RAF DNA mutation in serum for monitoring melanoma patients receiving biochemotherapy. Clin Cancer Res. 2007;13(7):2068–74.
Bettegowda C, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24.
Esposito A, et al. Monitoring tumor-derived cell-free DNA in patients with solid tumors: clinical perspectives and research opportunities. Cancer Treat Rev. 2014;40(5):648–55.
Gahan PB, Swaminathan R. Circulating nucleic acids in plasma and serum. Recent developments. Ann N Y Acad Sci. 2008;1137:1–6.
Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426–37.
Anker P, et al. Circulating nucleic acids in plasma or serum. Clin Chim Acta. 2001;313(1–2):143–6.
Allen D, et al. Role of cell-free plasma DNA as a diagnostic marker for prostate cancer. Ann N Y Acad Sci. 2004;1022:76–80.
Chun FK, et al. Circulating tumour-associated plasma DNA represents an independent and informative predictor of prostate cancer. BJU Int. 2006;98(3):544–8.
Sunami E, et al. Quantification of LINE1 in circulating DNA as a molecular biomarker of breast cancer. Ann N Y Acad Sci. 2008;1137:171–4.
Schwarzenbach H, et al. Detection and monitoring of cell-free DNA in blood of patients with colorectal cancer. Ann N Y Acad Sci. 2008;1137:190–6.
Diehl F, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–90.
Umetani N, et al. Prediction of breast tumor progression by integrity of free circulating DNA in serum. J Clin Oncol. 2006;24(26):4270–6.
Fleischhacker M, et al. Methods for isolation of cell-free plasma DNA strongly affect DNA yield. Clin Chim Acta. 2011;412(23–24):2085–8.
Slatko BE, Hiraizumi Y. Mutation induction in the male recombination strains of Drosophila melanogaster. Genetics. 1973;75(4):643–9.
Sonnenberg A, et al. Dielectrophoretic isolation and detection of cancer-related circulating cell-free DNA biomarkers from blood and plasma. Electrophoresis. 2014;35(12–13):1828–36.
Lam DC, et al. Plasma EGFR mutation detection associated with survival outcomes in advanced-stage lung cancer. Clin Lung Cancer. 2015;16(6):507–13.
Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–83.
Santiago-Dieppa DR, et al. Extracellular vesicles as a platform for ‘liquid biopsy’ in glioblastoma patients. Expert Rev Mol Diagn. 2014;14(7):819–25.
Yanez-Mo M, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066.
Andaloussi SEL, Mäger I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12(5):347–57.
van der Pol E, et al. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 2012;64(3):676–705.
Akers JC, et al. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J Neurooncol. 2013;113(1):1–11.
Minciacchi VR, Freeman MR, Di Vizio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin Cell Dev Biol. 2015;40:41–51.
Enderle D, et al. Characterization of RNA from exosomes and other extracellular vesicles isolated by a novel spin column-based method. PLoS One. 2015;10(8):e0136133.
Tauro BJ, et al. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56(2):293–304.
Schageman J, et al. The complete exosome workflow solution: from isolation to characterization of RNA cargo. Biomed Res Int. 2013;2013:253957.
van der Pol E, et al. Optical and non-optical methods for detection and characterization of microparticles and exosomes. J Thromb Haemost. 2010;8(12):2596–607.
Dragovic RA, et al. Sizing and phenotyping of cellular vesicles using nanoparticle tracking analysis. Nanomedicine. 2011;7(6):780–8.
Filipe V, Hawe A, Jiskoot W. Critical evaluation of nanoparticle tracking analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm Res. 2010;27(5):796–810.
Momen-Heravi F, et al. Alternative methods for characterization of extracellular vesicles. Front Physiol. 2012;3:354.
Garza-Licudine E, et al. Portable nanoparticle quantization using a resizable nanopore instrument—the IZON qNano. Conf Proc IEEE Eng Med Biol Soc. 2010;2010:5736–9.
de Vrij J, et al. Quantification of nanosized extracellular membrane vesicles with scanning ion occlusion sensing. Nanomedicine (Lond). 2013;8(9):1443–58.
van der Vlist EJ, et al. Fluorescent labeling of nano-sized vesicles released by cells and subsequent quantitative and qualitative analysis by high-resolution flow cytometry. Nat Protoc. 2012;7(7):1311–26.
Ueda K, et al. Antibody-coupled monolithic silica microtips for highthroughput molecular profiling of circulating exosomes. Sci Rep. 2014;4:6232.
Framework for Regulatory Oversight of Laboratory Developed Tests (LDTs) DRAFT GUIDANCE, F.a.D. Adminstration, Editor. 2014.
Clinical Laboratory Improvement Amendments of 1988. 1988, US Government Publishing Office.
Clinical Laboratory Improvement Amendments (CLIA). 4-16-14. Available from: http://www.fda.gov/MedicalDevices/DeviceRegulationandGuidance/IVDRegulatoryAssistance/ucm124105.htm. Accessed 3 Mar 2016.
Survey Procedures and Interpretive Guidelines for Laboratories and Laboratory Services. Available from: https://www.cms.gov/Regulations-and-Guidance/Legislation/CLIA/Interpretive_Guidelines_for_Laboratories.html. Accessed 7 Mar 2016.
Centers for Medicare & Medicaid Services. Clinical laboratory improvement amendments (CLIA). Available from: https://www.cms.gov/Regulations-and-Guidance/Legislation/CLIA/index.html. Accessed 7 Mar 2016.
HR 11124 An Act to amend the Federal Food, Drug, and Cosmetic Act to provide for the safety and effectiveness of medical devices intended for human use, and for other purposes. Library of Congress Thomas; 1976.
Draft Guidance for Industry, Food and Drug Administration Staff, and Clinical Laboratories 2014. Available from: http://www.fda.gov/downloads/medicaldevices/deviceregulationandguidance/guidancedocuments/ucm416685.pdf. Accessed 8 Mar 2016.
Ray T. Amid Competing LDT Regulatory Proposals, Common Ground but Key Disagreements for Congress to Consider. 2015. Available from: https://www.genomeweb.com/molecular-diagnostics/amid-competing-ldt-regulatory-proposals-common-ground-key-disagreements. Accessed 8 Mar 2016.
Acknowledgments
LS, LM and ML contributed to the writing of the manuscript. LS created the figure. ML and RV served as editors for the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
LS, LM, ML, and RV declare no conflicts of interest.
Funding
This work was funded by the National Institute of Health Grant # HHSN261201500035C and HHSN261201300034C.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Rights and permissions
About this article
Cite this article
Strotman, L.N., Millner, L.M., Valdes, R. et al. Liquid Biopsies in Oncology and the Current Regulatory Landscape. Mol Diagn Ther 20, 429–436 (2016). https://doi.org/10.1007/s40291-016-0220-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-016-0220-5