Abstract
Background and Objective
ABP 654 is a proposed biosimilar to ustekinumab reference product (RP), a human immunoglobulin isotype class G subclass 1 kappa monoclonal antibody that acts as an antagonist of interleukin (IL)-23 and IL-12. Ustekinumab RP is indicated for the treatment of some forms of plaque psoriasis, active psoriatic arthritis, Crohn’s disease, and ulcerative colitis. ABP 654 and ustekinumab RP utilize different expression systems, and the purpose of this study was to assess analytical similarity between ABP 654 and ustekinumab RP sourced from the United States (US) and the European Union (EU).
Methods
The analytical testing plan included general properties, primary structure, higher-order structure, product-related substances and impurities, particles and aggregates, biological activity, and thermal stability and degradation studies.
Results
ABP 654 was found to be analytically similar to ustekinumab RP with respect to physicochemical and biological properties, including structure, function, purity, and potency.
Conclusions
Based on a comprehensive similarity assessment, ABP 654 was found to be similar to ustekinumab RP, notwithstanding minor physicochemical differences that are not expected to have a clinically meaningful effect on safety or efficacy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
ABP 654 has been developed as a biosimilar product to the reference product Stelara® (ustekinumab), a biologic used for the treatment of Crohn's disease, ulcerative colitis, plaque psoriasis, and psoriatic arthritis. A comprehensive analytical similarity assessment has been carried out between ABP 654 and ustekinumab. Ustekinumab is expressed in an Sp2/0 cell line system, whereas ABP 654 is expressed in a Chinese hamster ovary cell line system glyco-engineered to enhance biosimilarity to ustekinumab. |
The totality of the data demonstrates that ABP 654 is analytically similar to ustekinumab; while there are some minor differences in ABP 654 compared with ustekinumab, these are not considered to be clinically meaningful. |
1 Introduction
ABP 654 is a proposed biosimilar to ustekinumab [1] reference product (RP), a human immunoglobulin (Ig) isotype class G subclass 1 kappa monoclonal antibody that binds to the common p40 subunit of cytokines interleukin (IL)-23 and IL-12. Binding of ustekinumab RP to IL-23 and IL-12 prevents interaction with their cognate receptors, resulting in modulation of immune system responses which play a key role in the pathophysiology of chronic inflammatory diseases [1,2,3]. In the United States (US) and the European Union (EU), ustekinumab RP is indicated for the treatment of some forms of plaque psoriasis, active psoriatic arthritis, Crohn’s disease, and ulcerative colitis [1, 4].
Ustekinumab RP is expressed in an Sp2/0 cell line system, whereas ABP 654 is expressed in a Chinese hamster ovary (CHO) cell line system glyco-engineered to enhance biosimilarity to ustekinumab RP. Recognizing that a change in expression system may result in some analytical differences, including variations in glycan profile, carefully considered cell line and manufacturing process development were applied, with emphasis on maintaining biological function critical to the mechanisms of action required for the clinical indications approved for ustekinumab RP, and minimizing any potential impact on safety, immunogenicity, or efficacy.
To gain regulatory approval of a biosimilar, US and EU guidelines recommend a stepwise totality-of-evidence (TOE) approach, which can be defined as the sum of data from comparative analytical, nonclinical, and clinical studies [5]. A TOE approach aims to demonstrate that there are no clinically meaningful differences in safety or efficacy between the biosimilar candidate and its RP [5,6,7,8,9,10].
Biosimilar development begins with an extensive analytical characterization of the RP, incorporating multiple orthogonal techniques to elucidate its structural and functional characteristics [11, 12]. The results from these studies are used to guide the development of a biosimilar candidate. An array of laboratory-based comparative assays is conducted to provide a comprehensive structural and functional profile of the molecule.
Consistent with US and EU guidelines [7, 10], this study was designed to assess biochemical, biophysical, and biological similarity, and to understand whether any differences between ABP 654 and ustekinumab RP have the potential to impact clinical performance. In these studies, ustekinumab RP was sourced from both the US and EU to satisfy the regulatory requirement, which states that the RP in these foundational comparative assessments must be locally sourced. This allows for the development of a scientific bridge such that only a single source of RP may be used in clinical evaluations.
2 Methods
Up to 22 lots of ABP 654 derived from unique drug substance lots were included in the assessment. Ustekinumab (US) and ustekinumab (EU) drug product lots were acquired on a regular basis and cover a period of more than 7 years. Up to 27 ustekinumab (US) and 40 ustekinumab (EU) lots were tested as part of the analytical similarity assessment. For attributes/activities where lot-to-lot variation is not expected and/or orthogonal methods were also utilized, a subset of lots was analyzed for similarity assessment. For ABP 654, clinical lots were included in the testing where a limited number of lots were used. For RP, lots available at the time of testing were selected. For attributes/activities where the data are quantitative in nature, the results of all unique ABP 654 lots were assessed against the minimum–maximum ranges established based on ustekinumab (US) and ustekinumab (EU) lots.
ABP 654 has the same dosage forms, presentations, and product strengths as the RP. ABP 654 is developed in multiple presentations, including 45 mg (0.5 mL) prefilled syringe (PFS), 90 mg (1 mL) PFS, 45 mg (0.5 mL) vial, and 130 mg (26 mL) vial. Since the drug substance manufacturing process is the same, results from these presentations were combined for similarity assessment, unless otherwise noted.
2.1 Analytical Assessments
2.1.1 Reduced Peptide Map
Reduced peptide map analysis was conducted by liquid chromatography with tandem mass spectrometry (LC–MS/MS) using a hybrid ion trap-Orbitrap mass spectrometer (Thermo Fisher Scientific). Samples were denatured with guanidine hydrochloride (HCl), then reduced with dithiothreitol (DTT; ThermoFisher) and alkylated using iodoacetic acid (Sigma). The desalted material was digested with trypsin (Roche Life Science) and the resulting cleavage fragments were separated by reversed-phase ultra-high-performance liquid chromatography (UHPLC) using a BEH C18 column (2.1 mm × 150 mm, 1.7 μm particle size; Waters) with an increasing gradient of 0.05 mM methionine, 0.1% trifluoroacetic acid (TFA) in acetonitrile. Eluting peaks were detected by ultraviolet (UV) absorbance (215 nm), and the peptides were introduced to the mass spectrometer (MS) by electrospray ionization to determine peptide masses.
2.1.2 Glycan Analysis
N-linked glycans were evaluated by glycan map analysis using hydrophilic interaction liquid chromatography (HILIC) with fluorescence detection (FD). N-glycans were released by treatment with the PNGase F (Agilent Technologies, Inc.; peptide-N4-[acetyl-β-glucosaminyl]-asparagine amidase EC 3.5.1.52). The reducing end of the released glycans was then labeled through reductive amination with a fluorescent tag, 2-aminobenzoic acid (Sigma-Aldrich), and the labeled glycans were separated using a Waters BEH Glycan column (2.1 × 150 mm, 1.7 μm particle size) in an acetonitrile mobile phase with an increasing gradient of ammonium formate in water. Peak identification was performed by coupling the UHPLC with an MS for verification against the expected glycan mass.
2.1.3 Sialic Acid Linkage Analysis
Samples were digested with Rapid PNGase F (New England Biolabs) in the presence of DTT. Released glycans were cleaned from buffer and residual protein using EB10 cartridges (QA-Bio). Dried glycans were labeled using 2-aminobenzamide acid (QA-Bio). Labeled glycans were purified from the reagent using Glycoclean S cartridges (Agilent). Dried labeled glycans were reconstituted in water. Released labeled glycans were treated each with neuraminidase Au (specific for 2-3,6,8,9 linkages; QA Bio) and neuraminidase Sp (specific for 2,3 linkages; QA-Bio). The samples were analyzed by HILIC-FD-MS as described above, except that a Waters BEH amide glycan HILIC column (2.1 × 100 mm, 1.7 μm particle size) was used. Glycans were assigned from their determined masses as well as by comparing retention times with those of available glycan standards.
2.1.4 Higher-Order Structure
For secondary structure assessment, the Fourier-transform infrared spectroscopy (FTIR) measurements of samples were performed side-by-side, without dilution, and at ambient temperature using an FTIR spectrometer. The spectrum of the buffer blank (ustekinumab drug product buffer) is recorded under identical conditions and is subtracted from the protein spectra. A single measurement is carried out for each sample. The second derivative spectrum is calculated using a 9-point smoothing of the normalized original spectra. The spectral similarity of each test lot against ustekinumab (US) and ustekinumab (EU) lots was quantitatively determined using the Thermo OMNIC software QC Compare function where 100% similarity indicates identical spectra [13].
For tertiary structure assessment, the near UV circular dichroism (CD) method measures the difference in absorption between left- and right-handed polarized light in the region of 240–340 nm. Samples were diluted before the near UV CD measurements. The near UV CD spectra of the samples were generated on a spectropolarimeter at ambient temperature using cuvettes with a pathlength of 1 cm. A single measurement was carried out for each sample. The spectra are corrected for small concentration differences and contributions from the buffer and are reported as mean residual molar ellipticity. The spectral similarity was quantitatively determined using the Thermo OMNIC software QC Compare function where 100% similarity indicates identical spectra [14].
The thermal stability of the samples was assessed side-by-side by differential scanning calorimetry (DSC) using a system in which temperature differences between the reference and sample cell are continuously measured and calibrated. The unfolding of the protein molecules appears as endothermic transitions on the DSC thermogram and can be characterized by the midpoints of the thermal transition temperatures (Tm). There are two thermal transitions (Tm1 and Tm2) corresponding to the unfolding of the fragment antigen binding and CH3 domains, respectively. Duplicate measurements were performed on all samples, and the results are reported as an average of the replicates.
2.1.5 Analytical Ultracentrifugation Sedimentation Velocity
Analytical ultracentrifugation sedimentation velocity (AUC-SV) was used to assess the hydrodynamic radius of ustekinumab RP and ABP 654. Samples were diluted before measurements by AUC-SV. The sedimentation velocity experiments were performed at 40,000 rpm, and the absorbance at 280 nm was recorded. Triplicate measurements were carried out for each sample and the mean results were reported as a percentage of monomer and high molecular weight (HMW) species. The HMW species were below the limit of quantitation for all samples.
2.1.6 Size‑Exclusion High‑Performance Liquid Chromatography with Light Scattering Detection
Size variants were assessed by size-exclusion high‑performance liquid chromatography with light scattering detection (SE-HPLC–LS), where SE-HPLC was coupled with a static light scattering detector, a refractive index (RI) detector, and a UV detector with wavelength set at 280 nm. Samples were injected without dilution into the SE-HPLC–LS system. For the molar mass calculation of eluting peaks, an RI increment value (ẟn/ẟc) of 0.182 mL/g was used.
2.1.7 Size Variants by Size-Exclusion Ultra-High-Performance Liquid Chromatography, Reduced and Non-Reduced Capillary Electrophoresis Sodium Dodecyl Sulfate
Native size variants were analyzed by size-exclusion ultra-high-performance liquid chromatography (SE-UHPLC). SE-UHPLC measurements were made on a UHPLC system with a BEH200 1.7 µm, 4.6 × 150 mm SEC column (Waters). Analytes were separated isocratically, monitored by UV absorbance at 280 nm, and purity was evaluated by determining the peak area of each species as a percentage of the total peak area. Capillary electrophoresis sodium dodecyl sulfate (CE-SDS) was used for separation of denatured protein size variants under reduced or non-reduced conditions. For the non-reduced condition, samples were denatured using SDS at 60 °C for 5 min. For the reduced condition, β-mercaptoethanol was added to the protein denaturation step to reduce the disulfide bonds and incubation was performed at 70 °C for 10 min. After denaturation, samples were injected into a bare fused silica capillary and separated based on hydrodynamic size resulting from an applied electric field in which the migration time of smaller size proteins is inversely related to overall size. Analytes were monitored by UV absorbance at 220 nm, and purity was evaluated by determining the corrected peak area of each species as a percentage of the total corrected peak area.
2.1.8 Charge Variants by Cation Exchange High-Performance Liquid Chromatography
Charged isoforms in samples were separated on a BioPro SP-F column (4.6 × 100 mm, 5 μm particle size; YMC). Eluted peaks using a salt gradient were monitored by UV absorbance at 280 nm and purity was evaluated by determining the peak area of each charged isoform group (acidic, main, and basic peaks) that eluted separately as a percentage of the total peak area. Samples were analyzed untreated, treated with carboxypeptidase B, and treated with carboxypeptidase B and PNGase F.
2.2 Functional Assessment
2.2.1 Inhibition of Interleukin (IL)-23- and IL-12-Mediated Signaling
IL-23 induces the signal transducer and activation of transcription (STAT) signaling pathway upon binding to the heterodimeric receptor comprised of IL-23R and IL-12Rβ1 chains. The ability of ABP 654 to inhibit IL-23-induced signaling was evaluated using a cell-based reporter assay. In this assay, IL-23R (HEK-293) cells containing IL-12Rβ1, IL-23R, and a sis-inducible element (SIE)-response element (RE) luciferase gene (Luc2P) were utilized to measure the activity of the IL-23-induced STAT signaling pathway. In the presence of IL-23, the cells were induced to generate a bioluminescent signal that can be detected with addition of luminescence reagent in a plate reader. The IL-23R reporter cells were incubated with a fixed concentration of IL-23 in combination with various concentrations of serially diluted sample. The reduction in the amount of luminescence detected was proportional to the degree of IL-23 blocking activity of the antibody. The test sample dose-response curve is fitted to a 4-parameter logistical equation and the relative activity of the sample was compared with that of the reference standard.
Binding of IL-12 to its heterodimeric receptor composed of IL-12Rβ2, and IL-12Rβ1 chains also induces the STAT signaling pathway. The principle for the inhibition of IL-12-induced signaling bioassay was the same as for the inhibition of IL-23-mediated signaling assay described above except that the IL-12R (HEK-293) cells containing IL-12Rβ1, IL-12Rβ2, and a SIE-RE-Luc2P is utilized to measure the IL-12-induced signaling.
2.2.2 IL-23 and IL-12 Receptor-Ligand Binding
Two receptor-ligand binding assays have been developed that use a bead-based amplified luminescent proximity homogeneous format that detects biomolecular interactions. For the IL-23 receptor-ligand binding assay, biotinylated human recombinant IL-12 receptor β1 (IL-12R-biotin) binds to histidine tagged IL-23 (IL-23-His). The format for the IL-12 receptor-ligand binding assay is the same as described for the IL-23 receptor-ligand binding assay except that IL-12 is used as the ligand. The donor beads are coated with a hydrogel that contains phthalocyanine, a photosensitizer, and streptavidin. The acceptor beads contain thioxene derivatives, as well as nickel chelate. When IL-12-R-biotin and IL-23-His or IL-12-His bind to each other, the acceptor and donor beads are brought into close proximity. When a laser is applied to this complex, ambient oxygen is converted to singlet oxygen by the donor beads and an energy transfer to the acceptor beads occurs, resulting in the production of luminescence that is measured in a plate reader. ABP 654 binds to IL-23-His/IL-12-His and prevents it from binding to the IL-12-R-biotin, thereby decreasing the luminescence output in a dose-dependent manner. The test sample dose-response curve is fitted to a 4-parameter logistical equation and the relative activity of the sample was compared with that of the reference standard. Each test sample response is compared with the reference standard.
2.2.3 Lack of Binding to Receptor-Bound IL-23 or IL-12 on the Cell Surface
Cells expressing the IL-23/IL-12 receptor, which are the same cells as used in the respective signaling assays, were pre-incubated with IL-23/IL-12 to allow association with the receptor. ABP 654 or ustekinumab RP were then added to examine lack of binding to the ligand-receptor complex using flow cytometric analysis. Phycoerythrin (PE)-labeled goat anti-mouse IgG or mouse anti-human IgG were added to detect antibody binding to pre-bound IL-23. A non-neutralizing antibody against the p40 subunit of IL-23/IL-12 was used as a positive control.
2.2.4 IL-23 and IL-12 Binding Kinetics and Affinity
Samples were tested for binding to IL-12 and IL-23 using a surface plasmon resonance (SPR) Biacore T200 optical biosensor. In brief, an anti-human Fc antibody was coupled to a flow cell and used to immobilize test samples. Recombinant human IL-23/IL-12 at various concentrations were injected over the captured antibody surfaces as well as a negative control reference surface. Kinetic binding data were globally fit using a 1:1 binding model to determine the association rate and dissociation rate constants (ka and kd), and the equilibrium dissociation binding constant (KD) for both IL-23 and IL-12.
2.2.5 Binding Specificity Against Cytokines Related to IL-23/IL-12
The IL-23/IL-12 binding specificity was characterized for ABP 654 and ustekinumab RP using a Biacore T200 optical biosensor. IL-23 cytokine family members IL-6, IL-35, and IL-39 were immobilized on a flow cell. The flow cells were injected with test samples and positive control antibodies anti-IL-6, anti-IL-35 (p35 subunit), and anti-IL-39 (p19 subunit), and assessed for reactivity.
2.2.6 Neonatal Fc Receptor Binding
Samples were tested using a bead-based amplified luminescent proximity homogeneous assay. The acceptor beads are coated with IgG. The donor beads are coated with a hydrogel that contains phthalocyanine, a photosensitizer, and streptavidin, which binds to biotinylated human neonatal Fc receptor (FcRn). When IgG is coated on the acceptor beads and binds to the biotinylated FcRn on the donor beads, it brings the acceptor and donor beads into close proximity. When ABP 654 is present at sufficient concentrations to inhibit the binding of IgG to the biotinylated FcRn at pH 6.0, a dose-dependent decrease in luminescence is observed. The test sample dose-response curve is fitted to a 4-parameter logistical equation and the relative activity of the sample was compared with that of the reference standard.
2.2.7 Fc Gamma Receptor (FcγR) Type Ia, IIa, IIb, IIIb Binding
The relative binding to Fc gamma receptor type Ia (FcγRIa), IIa (FcγRIIa), IIb (FcγRIIb), and IIIb (FcγRIIIb) was characterized by SPR using a Biacore T200 optical biosensor and an anti-histidine (anti-His) capture assay. In brief, an anti-His antibody was coupled to the flow cell, followed by the capture of recombinant human FcγRIa-His, FcγRIIa (131R)-His, FcγRIIb-His, and FcγRIIIb-His. Multiple sample concentrations were injected over the captured FcγR surfaces. Binding data were globally fit using the 1:1 binding model to determine the equilibrium KD for each concentration sample. The percentage relative affinity was determined by comparing the KD of each sample with that of the reference standard.
2.2.8 FcγR Type IIIa Binding
An SPR-based assay was used to evaluate the relative binding of FcγR type IIIa (FcγRIIIa) to the Fc domain of ABP 654 and ustekinumab RP using the high-affinity variant of FcγRIIIa, FcγRIIIa (158V). In this assay, FcγRIIIa was covalently immobilized on the sensor chip and samples were injected over the chip. Binding of ABP 654 or ustekinumab RP over a concentration range was measured. The relative binding was determined by measuring the binding response near the end of injection for each concentration of each sample dilution series, in duplicate. The test sample dose-response curve was fitted to a 4-parameter logistical equation and the relative activity of the sample was compared with that of the reference standard.
2.2.9 First Sub-Component of Complement Binding
A direct binding enzyme-linked immunosorbent assay (ELISA) method was developed to assess the binding of the Fc domain of the samples to the first sub-component of complement (C1q). Sample dilution series is adsorbed to a microtiter plate and probed with C1q. Bound C1q is detected with an anti-C1q-horseradish peroxidase (HRP) conjugated antibody. HRP activity is then detected using a colorimetric substrate, 3,3′,5,5′-tetramethylbenzidine, of the HRP enzyme. The reaction is stopped and the amount of colorimetric product is quantified by light absorbance. Each sample is compared with the reference standard.
2.2.10 Lack of Antibody-Dependent Cell-Mediated Cytotoxicity Activity
Antibody-dependent cell-mediated cytotoxicity (ADCC) activity was evaluated using the DiscoverX KiLR system, which employed U937 cells expressing membrane-associated IL-12 [15] as target cells and CD16a (FcγRIIIa) cytotoxic T-lymphocytes engineered to express CD16a (FcγRIIIa) as effector cells. ADCC activity was assessed by incubating interferon (IFN)-γ-stimulated U937 cells with multiple concentrations of test sample together with the KiLR CD16a effector cells at various effector-to-target cell ratios. A positive control antibody (anti-β2 microglobulin), which binds to the β2 microglobulin receptor on the surface of the U937 cells, was used as a positive control.
2.2.11 Lack of Complement-Dependent Cytotoxicity Activity
Complement-dependent cytotoxicity (CDC) activity was evaluated using U937 cells expressing membrane-associated IL-12 [15] as target cells and rabbit serum. CDC activity was determined by incubating IFN-γ-stimulated U937 cells with multiple concentrations of sample together with multiple concentrations of rabbit serum that provide a suitable level of target death. Changes in cell-based luminescence was used as a read out. A positive control consisting of a combination of anti-β2-microglobulin, anti-CD55, and anti-CD59 antibodies, which binds to the surface of U937 cells, was used to demonstrate CDC in the presence of rabbit serum.
2.2.12 Lack of Antibody-Dependent Cellular Phagocytosis Activity
Antibody-dependent cellular phagocytosis (ADCP) activity was evaluated using U937 cells expressing membrane-associated IL-12 [15] as target cells and human monocyte effector cells (CD14+, IL-2-stimulated). ADCP activity was determined by incubating IFN-γ-stimulated U937 cells with multiple concentrations of sample together with monocyte effector cells, at multiple effector-to-target cell ratios that would provide a suitable level of target death. A positive control molecule (anti-human leukocyte antigen [anti-HLA]), which binds to the surface of monocyte effector cells, was used to demonstrate ADCP in the presence of monocyte effector cells.
3 Results
The methods used for the analytical similarity assessment were selected based on knowledge regarding the structure, function, and heterogeneity of ABP 654 and ustekinumab RP, including those characteristics critical to the biological activity and stability of the product. The testing plan shown in Table 1 is organized into seven categories: general properties, primary structure, higher-order structure, product-related substances and impurities, particles and aggregates, biological activity, and thermal stability and degradation studies.
The methods were appropriately qualified to confirm they were fit for purpose. The results for select physicochemical assays are shown in Table 2, and the results for all biological activity assays are displayed in Table 3. Altogether, the results allow a meaningful and comprehensive assessment that increases the confidence that ABP 654 and ustekinumab RP are analytically similar.
3.1 Structural Properties
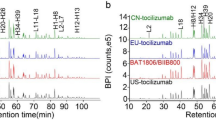
3.1.1 Comparison of Amino Acid Sequence by Reduced Peptide Map
Amino acid sequence was assessed using reduced peptide map analysis, including trypsin digestion of samples followed by LC–UV–MS/MS analysis. The observed tryptic peptide masses for ABP 654 and ustekinumab RP are as expected and within 100 ppm of their theoretical masses (data not shown). The tryptic peptide map chromatograms for the side-by-side comparison of ABP 654, ustekinumab (US), and ustekinumab (EU) are shown in Fig. 1. The peptide map profiles for both products are visually similar with minor differences primarily attributed to glycosylation profile and C-terminal lysine level differences due to the different cell lines used to manufacture ABP 654 and ustekinumab RP. However, the difference of CHO-derived N-acetylneuraminic acid (NANA)-containing glycans in ABP 654 and Sp2/0-derived N-glycolylneuraminic acid (NGNA)-containing glycans in ustekinumab RP is not expected to have clinical efficacy concerns as no difference in biological activity for the key mechanism of action of the three products was observed (Table 3), including fractions enriched for sialic acid-containing species from the three products (data not shown). Additionally, levels of identified post-translational modifications were compared between ABP 654 and ustekinumab RP. Overall, the observed chemical modification levels are very low and similar between ABP 654 and ustekinumab RP (data not shown). Moreover, ABP 654 has a lower level of C-terminal lysine compared with ustekinumab (EU) and is not considered clinically meaningful (see the Charge Variants section for further details).
3.1.2 Glycan Map and Sialic Acid Linkage
ABP 654 and ustekinumab RP are glycosylated IgG1 proteins, and each heavy chain (HC) of ABP 654 and ustekinumab RP contains 1 N-glycosylation site at N299 on the Fc domain. Ustekinumab RP is expressed in a murine mouse myeloma Sp2/0 cell line system [4], whereas ABP 654 is expressed in a CHO cell line system glyco-engineered to enhance biosimilarity to ustekinumab RP, with regard to the sialic acid linkages. A side-by-side glycan map analysis was performed comparing ABP 654 and ustekinumab RP. The glycan map chromatograms for ABP 654, ustekinumab (US), and ustekinumab (EU) lots are shown in Fig. 2. Identifications of individual glycans are shown in Fig. 2, and a glycan key is provided in Table 4. The most abundant glycan species in ABP 654, ustekinumab (EU), and ustekinumab (US) are bi-antennary complex structures with 0, 1, or 2 sialic acids (data not shown). The glycan map profiles of ABP 654 and ustekinumab RP are visually similar, with some new and missing species detected for ABP 654 compared with ustekinumab RP (Fig. 2), which is due to the differences in NANA-containing glycans for ABP 654 and NGNA-containing glycans for ustekinumab RP resulting from the difference in the production cell line. The different sialic acids, NANA versus NGNA, in ABP 654 and ustekinumab RP are not expected to impact clinical safety and efficacy.
Human serum IgGs contain NANA attached to a terminal galactose through an α-2,3 or α-2,6 linkage. CHO-derived glycoproteins exhibit complex glycan structures with terminal NANA through α2,3 linkages, while murine cell lines, such as Sp2/0, express glycoproteins with terminal NGNA through α2,6 linkages. The ABP 654-expressing CHO cell line was glyco-engineered to be similar to ustekinumab RP for the level of sialylation and type of linkage. To assess the sialic acid linkage type, ABP 654 and ustekinumab RP were subjected to a sialic acid linkage analysis. Figure 3a shows profiles of ABP 654 released N-glycans analyzed by HILIC (glycan map) for total glycans and sialidase-treated glycans. Among the two sialidases tested, neuraminidase Sp specifically removes sialic acid with α2,3 linkages, and neuraminidase Au removes sialic acid with all possible linkages (α2-3,6,8,9 linkages). We found that all ABP 654 sialyl glycan peaks are no longer detected after neuraminidase Au treatment, and only A2G2S2F (α2,3 and α2,6 linked) is no longer observed after neuraminidase Sp treatment, indicating all the other sialyl glycans in ABP 654 are α2,6 linked. Linkages of α2,8 and α2,9 only appear in glycans containing sialic acid-sialic acid motifs, which are not observed in the products analyzed in this work. Figure 3b, c show glycan profiles for total glycans and sialidase-treated glycans for ustekinumab (US and EU, respectively). Both products show very similar results, where all sialyl glycan peaks are no longer detected after neuraminidase Au treatment, and only A2G2Sg2Fa is no longer observed after neuraminidase Sp treatment, indicating all other sialyl glycans are α2,6 linked. Overall, these results confirm that the majority of the sialylated glycans for both ABP 654 and ustekinumab (US and EU) contain the α2,6-linked sialic acid.
Sialic acid linkage analysis of ABP 654 and ustekinumab (US and EU). Released N-glycans were analyzed by HILIC (glycan map) with and without treatment by linkage-specific neuraminidases. a HILIC profiles of ABP 654 for total glycans (black trace) and glycans after treatment with neuraminidase Au (specific for α2-3,6,8,9 linkages, blue trace) and neuraminidase Sp (specific for α2,3 linkage, pink trace). b HILIC profiles of ustekinumab (US) for total glycans (black trace) and glycans after treatment with neuraminidase Au (specific for α2-3,6,8,9 linkages, blue trace) and neuraminidase Sp (specific for α2,3 linkage, pink trace). c HILIC profiles of ustekinumab (EU) for total glycans (black trace) and glycans after treatment with neuraminidase Au (specific for α2-3,6,8,9 linkages, blue trace) and neuraminidase Sp (specific for α2,3 linkages, pink trace). EU European Union, HILIC hydrophilic interaction liquid chromatography, US United States
In total, glycan mapping by HILIC was performed on 22 ABP 654 lots, 27 ustekinumab (US) lots, and 40 ustekinumab (EU) lots. As part of the glycan map analysis, identified individual glycans were quantified and compiled into multiple glycan groups according to their potential biological relevancy for the similarity assessment.
The following glycan groups were evaluated as part of the similarity assessment, based on their potential to impact pharmacokinetics (PK) and/or effector functions: high mannose, sialylation, afucosylation, β-galactosylation, and α-galactosylation (Table 2). For high mannose glycans, small differences in levels (approximately 2%) [Table 2 and Online Resource 1) were observed between ABP 654 and ustekinumab RP; however, these minor differences did not have a clinically meaningful impact on PK based on clinical results [16]. ABP 654 was observed to have lower levels of afucosylation compared with ustekinumab RP (approximately 3%) [Table 2 and Online Resource 2]. The lower level of afucosylation in ABP 654 likely accounts for the lower FcγRIIIa binding activity (Table 3) but is not considered to have a clinically meaningful impact as ABP 654 and ustekinumab RP do not elicit effector functions (Table 3).
The three earliest ustekinumab (US) lots by expiration date were observed to have significantly lower levels of β-galactosylation and sialylation compared with all other US lots and all ustekinumab (EU) lots, which suggests a process change by the manufacturer. Thus, the minimum–maximum ranges for these glycan groups were significantly larger for the ustekinumab (US) lots compared with the ustekinumab (EU) lots. For β-galactosylation, the differences in levels were approximately 5% (Table 2 and Online Resource 3) for ABP 654 compared with ustekinumab (US and EU), not considering the first three US lots. The slightly higher level of β-galactosylation present in ABP 654 had no effect on biological activity, as the C1q-binding activity is similar and ustekinumab RP is shown to lack CDC activity (Table 3). Therefore, the difference in β-galactosylation is not expected to be clinically meaningful. For sialylation, the difference in levels was approximately 4% (Table 2 and Online Resource 4) between ABP 654 and ustekinumab (US and EU), not considering the first three US lots. The slightly higher level of sialylation present in ABP 654 is not considered clinically meaningful since no difference in biological activity for the key mechanism of action of the three products was observed (Table 3).
ABP 654 does not contain α-galactosylated glycans. Therefore, ABP 654 is not compared with ustekinumab RP for α-galactosylation. Given the low levels (≤ 5%) of α-galactosylated glycans seen in ustekinumab RP (Table 2 and Online Resource 5), the lack of α-galactosylation in ABP 654 is not considered to be clinically meaningful.
3.1.3 Size and Charge Variants
Product-related substances and impurities of ABP 654 and ustekinumab RP were assessed using a combination of methods that evaluated size and charge variants. Size variants were assessed by SE-UHPLC, reduced capillary electrophoresis sodium dodecyl sulfate (rCE-SDS), and non-reduced capillary electrophoresis sodium dodecyl sulfate (nrCE-SDS), and charge variants were assessed by cation exchange high-performance liquid chromatography (CEX-HPLC). This similarity assessment includes the PFS presentations, 45 mg vial presentation, and drug substance lots, since they share a common protein concentration and formulation; the 130 mg vial presentation is not included in this assessment since the different protein concentration and formulation impacted the size variant stability profiles (data not shown). While the changes in the drug product manufacturing process, container closure system, and formulation were not expected to have an impact on the charge variants, the 130 mg vial presentation is not included in this assessment for charge variants.
A side-by-side SE-UHPLC analysis was carried out on ABP 654 and ustekinumab RP, and the profile comparison is presented in Fig. 4a. The SE-UHPLC profiles are visually similar with no new or missing peaks observed for ABP 654 as compared with ustekinumab RP. In total, SE-UHPLC was performed on 22 ABP 654, 22 ustekinumab (US), and 33 ustekinumab (EU) lots. Generally, the ABP 654 lots are above the ustekinumab RP range for levels of HMW, but the difference is < 0.5% (Table 2). The majority of ABP 654 HMW species is in the form of dimer (SE-HPLC–LS results) [Table 2], which is not likely to be immunogenic. Additionally, no differences between ABP 654 and ustekinumab RP were observed in the biological activity related to the mechanism of action (Table 3). Therefore, these minor differences (< 0.5%) in levels of SE-UHPLC HMW between ABP 654 and ustekinumab are not considered clinically meaningful.
Size variant assessment of ABP 654 and ustekinumab (US and EU). a Side-by-side SE-UHPLC analysis. b Side-by-side rCE-SDS analysis. c Side-by-side nrCE-SDS analysis. EU European Union, HC heavy chain, HMW high molecular weight, LC liquid chromatography, LMW low molecular weight, MMW mid molecular weight, NGHC non-glycosylated heavy chain, US United States
A side-by-side rCE-SDS analysis was carried out on ABP 654 and ustekinumab RP, and the profile comparison is presented in Fig. 4b. The rCE-SDS profiles are visually similar across the two products with no new or missing peaks observed at >0.5% for ABP 654 as compared with ustekinumab RP. In total, rCE-SDS was performed on 22 ABP 654, 22 ustekinumab (US), and 33 ustekinumab (EU) lots. Generally, minor differences (< 1%) were observed in the HC + LC, fragments, and non-glycosylated HC (NGHC) levels between ABP 654 and ustekinumab (Table 2). The slightly lower levels of HC + LC in ABP 654 is primarily due to slightly higher levels of NGHC. These minor differences do not impact biological function related to the mechanism of action (Table 3) and therefore are not considered clinically meaningful.
A side-by-side nrCE-SDS analysis was carried out on ABP 654 and ustekinumab RP, and the profile comparison is presented in Fig. 4c. The nrCE-SDS profiles are visually similar across the two products with no new or missing peaks observed for ABP 654 as compared with ustekinumab RP. In total, nrCE-SDS was performed on 22 ABP 654, 22 ustekinumab (US), and 33 ustekinumab (EU) lots. Minor differences were observed in the main peak and pre-peak levels between ABP 654 and ustekinumab RP. The slightly lower levels of main peak in ABP 654 are primarily due to slightly higher levels (< 2% difference) of pre-peaks (Table 2). Most pre-peak species represent properly assembled antibodies with one or more broken interchain disulfide bonds [17]. These species are known to convert in redox environments, such as those present in physiological conditions, back to fully disulfide-linked antibodies [18]. In addition, this minor difference in pre-peaks has no impact on the biological activity related to the mechanism of action (Table 3) and is not considered clinically meaningful.
A side-by-side CEX-HPLC analysis was carried out on ABP 654 and ustekinumab RP, and the profile comparison is presented in Fig. 5a. The CEX-HPLC profiles are visually similar in the acidic and main peak regions and ABP 654 shows a lower level of basic peaks primarily resulting from differences in levels of C-terminal lysine (see below); however, no new peaks were observed for all ABP 654 lots compared with ustekinumab RP. In total, CEX-HPLC was performed on 22 ABP 654, 22 ustekinumab (US), and 33 ustekinumab (EU) lots. ABP 654 has a lower level of basic peaks compared with ustekinumab RP, due to a lower level of unprocessed HC C-terminal lysine variants. Additionally, ABP 654 contains a higher level of acidic peaks compared with ustekinumab RP, due to a lower level of basic peaks and slightly higher level of sialic acid-containing species in ABP 654 (Table 2). However, the differences between ABP 654 and ustekinumab RP in the levels of acidic and basic peaks are not considered clinically meaningful, based on charge variant fraction collection and biological activity assessment (data not shown).
The charge variants were further characterized by enzymatic treatment of the two products with carboxypeptidase B, with or without PNGase F, followed by CEX-HPLC analysis. Upon treatment with carboxypeptidase B, the levels of basic peaks between ABP 654 and ustekinumab (US and EU) are similar (Fig. 5b) with small differences in acidic peaks. The results demonstrate that the primary contributor to the differences in CEX-HPLC basic peak levels of untreated samples is due to lower levels of C-terminal lysine in ABP 654. Differences in acidic species after treatment with carboxypeptidase B is considered to be mostly due to charged (sialic acid-containing) glycans. As shown in Fig. 5c, upon treatment with PNGase F, the carboxypeptidase B-treated ABP 654 samples showed a similar level of acidic species compared with the equivalently treated ustekinumab RP samples.
Overall, small differences between ABP 654 and ustekinumab (US and EU) in the levels of size and charge variants, as well as glycans (Sect. 3.1.2), did not adversely impact comparative clinical studies in terms of PK [16], safety, and efficacy (data not shown). This finding is similar to the observation that several European Medicines Agency-authorized biosimilars had small analytical differences compared with the RP that did not result in clinically meaningful differences [19].
3.2 Biological Activity
The primary mechanism of action of ABP 654 and ustekinumab RP is through disruption of IL-23- and IL-12-mediated signaling and cytokine cascades, resulting in suppression of inflammation. The assays included in the biological activity similarity assessment were selected to interrogate a wide range of biological functions to assure that any minor biochemical variations would have no significant impact on the biological performance of ABP 654 as compared with ustekinumab RP.
Inhibition of IL-23-mediated signaling is a key component of the primary mechanism of action for ABP 654. A cell-based bioassay was used to assess the ability of ABP 654 and ustekinumab RP to inhibit the binding of IL-23 to its heterodimeric receptor, preventing the induction of the STAT signaling pathway. This method was performed on 22 ABP 654, 16 ustekinumab (US), and 31 ustekinumab (EU) lots. The range of ABP 654 and ustekinumab RP for relative percentage inhibition are shown in Table 3. The individual value plots for each product are displayed in Fig. 6a, and these results demonstrate that ABP 654 and ustekinumab RP are similar for inhibition of IL-23-mediated signaling.
IL-23 receptor-ligand binding is a key proximal component of the primary mechanism of action for ABP 654. The IL-23 receptor-ligand binding by bead-based luminescence proximity assay was performed on 22 ABP 654, 16 ustekinumab (US), and 31 ustekinumab (EU) lots. The range of ABP 654 and ustekinumab RP for relative percentage binding is shown in Table 3. The individual value plots for each product are displayed in Fig. 6b. These results demonstrate that ABP 654 and ustekinumab RP are similar for binding to IL-23.
Inhibition of IL-12-mediated signaling is also a key component of the primary mechanism of action for ABP 654. A cell-based bioassay was used to assess the ability of ABP 654 and ustekinumab RP to inhibit the binding of IL-12 to its heterodimeric receptor, preventing the induction of the STAT signaling pathway. This method was performed on 22 ABP 654, 15 ustekinumab (US), and 31 ustekinumab (EU) lots. The range of ABP 654 and ustekinumab RP for relative percentage inhibition is shown in Table 3. The individual value plots for each product are displayed in Fig. 6c. These results demonstrate that ABP 654 and ustekinumab RP are similar for inhibition of IL-12-mediated signaling.
IL-12 receptor-ligand binding is a key proximal component of the primary mechanism of action for ABP 654. The IL-12 receptor-ligand binding by bead-based luminescence proximity assay was performed on 22 ABP 654, 15 ustekinumab (US), and 31 ustekinumab (EU) lots. The range of ABP 654 and ustekinumab RP for relative percentage binding is shown in Table 3. The individual value plots for each product are displayed in Fig. 6d, and these results demonstrate that ABP 654 and ustekinumab RP are similar for binding to IL-12.
4 Discussion
This study was designed to evaluate the analytical similarity of the proposed biosimilar ABP 654 to ustekinumab (US) and ustekinumab (EU). A comprehensive set of complementary methods was used, including general properties, primary structure, higher-order structure, product-related substances and impurities, particles and aggregates, biological activity, and thermal stability and degradation studies. There were some minor differences in physiochemical attributes observed between ABP 654 and ustekinumab RP, primarily due to the different cell lines used between ABP 654 and ustekinumab RP. However, results of the biological and functional assessments showed that these minor physicochemical differences do not affect biological functions relevant to the mechanism of action of ABP 654 and ustekinumab RP. The results also demonstrated that ustekinumab sourced from both the US and EU were analytically similar.
Results from reduced peptide mapping demonstrated that the amino acid sequence of ABP 654 was identical to that of ustekinumab RP. Chemical modifications were similar between ABP 654 and ustekinumab RP. Differences were observed for C-terminal lysine levels and for sialic acid-containing glycans, CHO-derived NANA-containing glycans in ABP 654 and Sp2/0-derived NGNA, and α-gal-containing glycans in ustekinumab RP; however, these differences are not expected to pose clinical safety or efficacy concerns. Small differences in glycan groups (high mannose, sialylation, afucosylation, β-galactosylation, and α-galactosylation) were observed between ABP 654 and ustekinumab RP. Altogether, the differences in glycan types and levels of glycan groups are not considered meaningful due to a lack of observable differences in biological activity for the key mechanism of action of the three products (Table 3), including fractions enriched in sialic acid-containing species. Moreover, results from comparative clinical studies demonstrated similarity of ABP 654 with ustekinumab RP in terms of PK [16], safety, and efficacy (data not shown).
Product-related substances and impurities were assessed by methods that analyze size and charge variants. Size variants for ABP 654 and ustekinumab RP were considered similar, notwithstanding minor differences that are not considered clinically meaningful. Charge variants for ABP 654 and ustekinumab RP were considered similar after excluding C-terminal lysine and glycans from the analyses. These differences are not expected to be clinically meaningful, as detailed above.
While ustekinumab RP binds to FcγR(s) and C1q, it is not expected to induce ADCC, ADCP, or CDC, since IL-23 and IL-12 primarily exist as soluble secreted proteins. Additionally, ustekinumab RP does not bind to IL-23 and IL-12 that is already bound to cell surface receptors, and there is no evidence that antibody Fc-mediated effector functions that result in cellular depletion (such as ADCC, ADCP, or CDC) occur or contribute to the mechanism of action of ustekinumab [1, 4, 20]. Based on comparative assessments of the biological activity of ABP 654 and ustekinumab RP, all biological activities relevant to the primary mechanism of action, including inhibition of IL-23-mediated signaling, inhibition of IL-12-mediated signaling, IL-23 receptor ligand binding, and IL-12 receptor ligand binding were found to be similar.
5 Conclusions
The purpose of this study was to assess analytical and functional similarity between the proposed biosimilar ABP 654 and ustekinumab RP sourced from the US and EU. Based on a comprehensive similarity assessment, ABP 654 was found to be analytically similar to ustekinumab RP with respect to physicochemical and biological properties, including structure, function, purity, and potency. The results also demonstrate that ustekinumab (US) is analytically similar to ustekinumab (EU). While there were some minor differences in ABP 654 compared with ustekinumab RP, these were not considered to be clinically meaningful and are unlikely to have an impact on PK, efficacy, or safety.
References
STELARA® (ustekinumab). United States prescribing information. Horsham: Janssen Biotech, Inc.; 2020.
Ryan C, Thrash B, Warren RB, Menter A. The use of ustekinumab in autoimmune disease. Expert Opin Biol Ther. 2010;10(4):587–604. https://doi.org/10.1517/14712591003724670.
Teng MW, Bowman EP, McElwee JJ, Smyth MJ, Casanova JL, Cooper AM, et al. IL-12 and IL-23 cytokines: from discovery to targeted therapies for immune-mediated inflammatory diseases. Nat Med. 2015;21(7):719–29. https://doi.org/10.1038/nm.3895.
European Medicines Agency. EPAR ustekinumab (Stelara), 2019. Available at: https://www.ema.europa.eu/en/medicines/human/EPAR/stelara. Accessed 5 Dec 2022.
Markus R, Liu J, Ramchandani M, Landa D, Born T, Kaur P. Developing the totality of evidence for biosimilars: regulatory considerations and building confidence for the healthcare community. BioDrugs. 2017;31(3):175–87. https://doi.org/10.1007/s40259-017-0218-5.
US Food and Drug Administration. Quality considerations in demonstrating biosimilarity of a therapeutic protein product to a reference product guidance for industry. April 2015. Available at: https://www.fda.gov/media/135612/download. Accessed 5 Dec 2022.
European Medicines Agency. Guideline on similar biological medicinal products. 23 October 2014. CHMP/437/04 Rev. 1. Available at: http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2014/10/WC500176768.pdf. Accessed 5 Dec 2022.
US Food and Drug Administration. Guidance for industry: scientific considerations in demonstrating biosimilarity to a reference product. April 2015. Available at: https://www.fda.gov/media/82647/download. Accessed 5 Dec 2022.
Kim H, Alten R, Avedano L, Dignass A, Gomollon F, Greveson K, et al. The future of biosimilars: maximizing benefits across immune-mediated inflammatory diseases. Drugs. 2020;80(2):99–113. https://doi.org/10.1007/s40265-020-01256-5.
US Food and Drug Administration. Development of therapeutic protein biosimilars: comparative analytical assessment and other quality-related considerations guidance for industry. May 2019. Available at: https://www.fda.gov/media/159261/download. Accessed 5 Dec 2022.
Abraham I, Sun D, Bagalagel A, Altyar A, Mohammed A, Tharmarajah S, et al. Biosimilars in 3D: definition, development and differentiation. Bioengineered. 2013;4(4):203–6. https://doi.org/10.4161/bioe.25067.
McCamish M, Woollett G. Worldwide experience with biosimilar development. MAbs. 2011;3(2):209–17. https://doi.org/10.4161/mabs.3.2.15005.
Jiang Y, Li C, Nguyen X, Muzammil S, Towers E, Gabrielson J, et al. Qualification of FTIR spectroscopic method for protein secondary structural analysis. J Pharm Sci. 2011;100(11):4631–41. https://doi.org/10.1002/jps.22686.
Li CH, Nguyen X, Narhi L, Chemmalil L, Towers E, Muzammil S, et al. Applications of circular dichroism (CD) for structural analysis of proteins: qualification of near- and far-UV CD for protein higher order structural analysis. J Pharm Sci. 2011;100(11):4642–54. https://doi.org/10.1002/jps.22695.
Fan X, Sibalic V, Niederer E, Wuthrich RP. The proinflammatory cytokine interleukin-12 occurs as a cell membrane-bound form on macrophages. Biochem Biophys Res Commun. 1996;225(3):1063–7. https://doi.org/10.1006/bbrc.1996.1295.
Chow V, Mytych DT, Das S, Franklin J. Pharmacokinetic similarity of ABP 654, an ustekinumab biosimilar candidate: results from a randomized, double-blind study in healthy subjects. Clin Pharmacol Drug Dev. 2023;12(9):863–873. https://accp1.onlinelibrary.wiley.com/doi/10.1002/cpdd.
Hutterer KM, Hong RW, Lull J, Zhao X, Wang T, Pei R, et al. Monoclonal antibody disulfide reduction during manufacturing: Untangling process effects from product effects. MAbs. 2013;5(4):608–13. https://doi.org/10.4161/mabs.24725.
Wang T, Liu YD, Cai B, Huang G, Flynn GC. Investigation of antibody disulfide reduction and re-oxidation and impact to biological activities. J Pharm Biomed Anal. 2015;102:519–28. https://doi.org/10.1016/j.jpba.2014.10.023.
Guillen E, Ekman N, Barry S, Weise M, Wolff-Holz E. A data driven approach to support tailored clinical programs for biosimilar monoclonal antibodies. Clin Pharmacol Ther. 2023;113(1):108–23. https://doi.org/10.1002/cpt.2785.
Benson JM, Peritt D, Scallon BJ, Heavner GA, Shealy DJ, Giles-Komar JM, et al. Discovery and mechanism of ustekinumab: a human monoclonal antibody targeting interleukin-12 and interleukin-23 for treatment of immune-mediated disorders. MAbs. 2011;3(6):535–45. https://doi.org/10.4161/mabs.3.6.17815.
Acknowledgements
The authors thank Kristin Abrams, Jung Choi, Jill Crouse-Zeineddini, Trupti Guhan, Nany Jiao, Palanisamy Kanakaraj, Sheeba Kazi, Bin Lu, Alexis Lueras, and Chris Rollins for scientific and experimental support. Medical writing assistance was provided by Alex Romero, PhD (Amgen Inc.), under the direction of Sonya Lehto, PhD (Amgen Inc.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was funded by Amgen Inc., Thousand Oaks, CA, USA.
Conflict of interest
Greg Cantin, Qian Liu, Bhavana Shah, Scott Kuhns, Mats Wikström, Shawn Cao, and Jennifer Liu are employees and stockholders of Amgen Inc.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication, availability of data, and material code availability
There is a plan to share data. This may include de-identified individual patient data for variables necessary to address the specific research question in an approved data-sharing request, and also related data dictionaries, study protocol, statistical analysis plan, informed consent form, and/or clinical study report. Data sharing requests relating to data in this manuscript will be considered after the publication date and (1) this product and indication (or other new use) have been granted marketing authorization in both the US and Europe, or (2) clinical development discontinues and the data will not be submitted to regulatory authorities. There is no end date for eligibility to submit a data sharing request for these data. Qualified researchers may submit a request containing the research objectives, the Amgen product(s) and Amgen study/studies in scope, endpoints/outcomes of interest, statistical analysis plan, data requirements, publication plan, and qualifications of the researcher(s). Complete details are available at https://wwwext.amgen.com/science/clinical-trials/clinical-data-transparency-practices/clinical-trial-data-sharing-request/. In general, Amgen does not grant external requests for individual patient data for the purpose of re-evaluating safety and efficacy issues already addressed in the product labeling. A committee of internal advisors reviews requests. If not approved, a Data Sharing Independent Review Panel may arbitrate and make the final decision. Requests that pose a potential conflict of interest or an actual or potential competitive risk may be declined at Amgen’s sole discretion and without further arbitration. Upon approval, information necessary to address the research question will be provided under the terms of a data sharing agreement. This may include anonymized individual patient data and/or available supporting documents, containing fragments of analysis code where provided in analysis specifications. Further details are available at http://www.amgen.com/datasharing.
Author contributions
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Supplementary Information
Below is the link to the electronic supplementary material.





Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Cantin, G., Liu, Q., Shah, B. et al. Analytical and Functional Similarity of the Biosimilar Candidate ABP 654 to Ustekinumab Reference Product. Drugs R D 23, 421–438 (2023). https://doi.org/10.1007/s40268-023-00441-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40268-023-00441-7