Abstract
Introduction
Recently, increasing FcRn binding by Fc engineering has become a promising approach for prolonging the half-life of therapeutic monoclonal antibodies (mAbs). This study is the first to investigate the optimization of an allometric scaling approach for engineered mAbs based on cynomolgus monkey data to predict human pharmacokinetics.
Methods
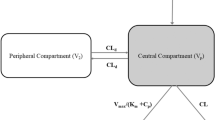
Linear two-compartmental model parameters (clearance [CL]; volume of distribution in the central compartment [Vc]; inter-compartmental clearance [Q]; volume of distribution in the peripheral compartment [Vp]) after the intravenous (IV) injection of engineered mAbs (M252Y/S254T/T256E or M428L/N434S mutations) in cynomolgus monkeys and humans were collected from published data. We explored the optimal exponent for allometric scaling to predict parameters in humans based on cynomolgus monkey data. Moreover, the plasma concentration–time profile of engineered mAbs after IV injection in humans was predicted using parameters estimated based on an optimized exponent.
Results
For engineered mAbs, a significant positive correlation between cynomolgus monkeys and humans was observed for CL, but not for other parameters. Whereas conventional exponents (CL: 0.8, Q: 0.75, Vc: 1.0, Vp: 0.95) previously established for normal mAbs showed poor prediction accuracy for CL and Q of engineered mAbs, the newly optimized exponents (CL: 0.55, Q: 0.6, Vc: 0.95, Vp: 0.95) achieved superior predictability for engineered mAbs. Moreover, the optimized exponents accurately predicted plasma mAb concentration–time profiles after IV injection of engineered mAbs in humans.
Conclusions
We found that engineered mAbs require specially optimized exponents to accurately predict pharmacokinetic parameters and plasma concentration–time profiles after IV injections in humans based on cynomolgus monkey data. This optimized approach can contribute to a more accurate prediction of human pharmacokinetics in the development of engineered mAbs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
It was demonstrated that the allometric scaling approach with conventional exponents is not appropriate for predicting two-compartment model parameters of engineered monoclonal antibodies (mAbs) in humans. |
An allometric scaling approach with optimized exponents was developed that showed good prediction accuracy for two-compartment model parameters of engineered mAbs in humans based on cynomolgus monkey data. |
Plasma concentration–time profiles of engineered mAbs after IV injection in humans were accurately predicted using the allometric scaling approach with optimized exponents. |
1 Introduction
Therapeutic monoclonal antibodies (mAbs) are a promising therapeutic modality for the treatment of several diseases. Over 100 mAbs have been approved by the US FDA and over 800 mAbs are currently in the clinical stage of development [1]. Moreover, several unique engineered mAbs—such as bispecific antibodies [2], recycling/sweeping antibodies [3, 4], local activable antibodies [5], and antibody–drug conjugates (ADC) [6]—have been developed. Significant advantages of mAbs over other therapeutic modalities are high/selective target antigen binding and a long half-life in the body. The long half-life is mainly achieved by their large molecular weight and neonatal Fc receptor (FcRn)-mediated endosomal recycling. Large molecular weight limits glomerular filtration and FcRn-mediated endosomal recycling prevents lysosomal degradation, giving a longer half-life than other therapeutic modalities.
While mAbs already have a long half-life in the body, several potential ways to extend it even further have been investigated. One approach is reducing the isoelectric point (pI) or increasing negative charge through mutagenesis. The cell surface is negatively charged, and mAbs with low pI/negative charge are repelled by negatively charged cell surfaces, which slows the endocytosis rate and prolongs half-life [7, 8]. Another approach is improving FcRn-mediated endosomal recycling efficiency though mutagenesis. Dall'Acqua et al. found that M252Y/S254T/T256E (YTE) mutations showed 10-fold increased binding to human and cynomolgus monkey FcRn at pH 6.0 [9]. They demonstrated that YTE mutations successfully increased the half-life by 4-fold in cynomolgus monkeys. Also, Zalevsky et al. found that M428L/N434S (LS) mutations showed an 11-fold increase in binding to human FcRn at pH 6.0 and a 3.2-fold improvement of half-life in cynomolgus monkeys [10]. Furthermore, both YTE and LS mutations have been reported to successfully prolong the half-life of mAbs in humans without increasing immunogenicity risk [11,12,13]. Since it has already been proven in several clinical trials that pharmacokinetics of mAbs can be improved by mutations, the application of these mutations in the pharmaceutical industry will increase.
In drug development, it is important to predict human pharmacokinetics in the preclinical stage. For mAbs, linear pharmacokinetics in humans was reported to be accurately predicted by an allometric scaling approach from cynomolgus monkeys [14, 15]. Our group established optimal exponents for predicting linear two-compartmental model parameters (clearance [CL]; volume of distribution in the central compartment [Vc]; inter-compartmental clearance [Q]; volume of distribution in the peripheral compartment [Vp]) in humans from cynomolgus monkeys using an allometric scaling approach [16]. This approach successfully predicted plasma mAb concentration–time profiles after intravenous (IV) injection in humans for 23 mAbs. Although this allometric scaling approach can be widely used for the prediction of human pharmacokinetics in mAbs development, it was only validated using mAbs without mutations that increase FcRn binding. Therefore, its applicability to engineered mAbs is uncertain.
In this study, first, we investigated whether the conventional allometric scaling approach could be used to predict human pharmacokinetics for engineered mAbs. Then, we explored optimal exponents in the allometric scaling approach for engineered mAbs using reported pharmacokinetic data from cynomolgus monkeys and humans. This is the first report to comprehensively validate the methodology for predicting the pharmacokinetics of engineered mAbs in humans.
2 Materials and Methods
2.1 Data Collection
Linear two-compartment model parameters (CL, Q, Vc, and Vp) and plasma mAb concentration–time profiles of engineered mAbs with YTE or LS mutations after IV injection in cynomolgus monkey and humans were obtained from published data. Additionally, information on the immunoglobulin G (IgG) subclass and target antigen was also collected. To exclude the effect of target-mediated drug disposition (TMDD) on analysis, only mAbs showing linear pharmacokinetics without TMDD in both cynomolgus monkeys and humans were selected for analysis. However, it should be noted that the possibility of involvement of TMDD cannot be completely excluded. When body weight information was unavailable in published data, 3 kg for cynomolgus monkeys and 75 kg for humans were applied for analysis. When CL, Q, Vc, and Vp were available in published data, these values were used for analysis. If CL, Q, Vc, and Vp were unavailable, parameters were estimated using a two-compartment model from plasma mAb concentration–time profiles. These profiles were obtained by scanning figures from published data using UnGraph 5 (Biosoft). Linear two-compartment model parameters for a total of nine engineered mAbs were collected. However, among the nine engineered mAbs, the plasma mAb concentration–time profiles after IV injection in humans for tixagevimab/AZD8895 and cilgavimab/AZD1061 were unavailable in published data. Therefore, having excluded these two, a total of seven engineered mAbs were used for the analysis of plasma mAb concentration–time profiles.
2.2 Allometric Scaling Approach with Conventional Exponents
Our previous study established an allometric scaling approach with conventional exponents for normal mAbs [16]. Linear two-compartment model parameters of nine engineered mAbs in humans were predicted by the allometric scaling approach with conventional exponents as shown in the following equations.
BW and e represent body weight (/kg) and exponent. Units in the above equations were mL/day for CL and Q and mL for Vc, and Vp. Conventional exponents for CL, Q, Vc, and Vp in the allometric scaling approach were 0.8, 0.75, 1.0, and 0.95, respectively.
2.3 Optimization of Exponents for Engineered mAbs
To establish an optimized allometric scaling approach for engineered mAbs from cynomolgus monkey to humans, optimal exponents for CL, Q, Vc, and Vp were explored. In the allometric scaling approach equation as shown in Sect. 2.2, exponents were examined in 0.05 increments from 0.5 to 0.8 for CL and Q and from 0.8 to 1.1 for Vc and Vp. Then, prediction accuracy for each exponent was evaluated by comparing predicted values with observed values.
2.4 Prediction of Plasma mAb Concentration–Time Profiles After IV Injection in Humans
Next, to evaluate the utility of optimal exponents, the plasma mAb concentration–time profiles of engineered mAbs after IV injection in humans were predicted from cynomolgus monkey data using allometric scaling approaches with both conventional and optimized exponents. The plasma mAb concentration–time profiles in humans predicted by the two approaches were compared with observed values.
2.5 Analysis
All fittings and simulations for plasma mAb concentration–time profiles were performed using SAAMII software (The Epsilon Group, Charlottesville, VA, USA). Relative weight (1/y^2) was used in all fittings. All figures and statistical analyses were prepared using GraphPad Prism 9 (GraphPad Software, San Diego, CA, USA). Correlation of linear two-compartment model parameters between cynomolgus monkeys and humans was statistically analyzed. Pearson correlation coefficient r value was significant when p < 0.05.
3 Results
3.1 Pharmacokinetic Parameters of Engineered mAbs in Cynomolgus Monkeys and Humans
A total of five mAbs with YTE mutations and four mAbs with LS mutations were selected for analysis (Table 1). All mAbs were reported to show linear pharmacokinetics in cynomolgus monkeys and humans. The IgG subclass of all mAbs was IgG1. The geometric means of CL for nine engineered mAbs in cynomolgus monkeys and humans were 3.86 and 0.91 mL/day/kg, respectively. These values were significantly lower than those previously reported in normal mAbs (6.14 mL/day/kg in cynomolgus monkeys and 3.32 mL/day/kg in humans) [16], demonstrating the effect of mutations to increase FcRn binding in both cynomolgus monkeys and humans. On the other hand, Vc and Vp for engineered mAbs were similar to those for normal mAbs in both cynomolgus monkey and humans as previously reported [16]. Although Q in cynomolgus monkeys was similar between normal mAbs and engineered mAbs, Q in humans for engineered mAbs was slightly lower than that for normal mAbs. While there was a significant positive correlation of CL between cynomolgus monkeys and humans (p < 0.05, r = 0.81), no significant correlation of Q, Vc, and Vp was observed.
3.2 Prediction of Two-Compartment Model Parameters of Engineered mAbs in Humans
First, two-compartment model parameters (CL, Q, Vc, and Vp) of nine engineered mAbs in humans were predicted from that in cynomolgus monkeys using the allometric scaling approach with conventional exponents. As shown in Fig. 1, although Vc and Vp were reasonably predicted using exponents of 1.0 and 0.95 within a 1.5-fold difference of observed values (78% for Vc and 89% for Vp within 1.5-fold of observed values), CL and Q showed a clear tendency to be over-predicted using exponents of 0.8 and 0.75 (11% for CL and 56% for Q within 1.5-fold of observed values). This result suggests that the translational rule of pharmacokinetics from cynomolgus monkeys to humans between normal mAbs and engineered mAbs was different. Therefore, to establish an optimized allometric scaling approach for engineered mAbs, optimal exponents for CL, Q, Vc, and Vp were explored by assessing the predictability of each parameter as shown in Fig. 2. As a result, the optimal exponents for CL, Q, Vc, and Vp were estimated to be 0.55, 0.6, 0.95, and 0.95, respectively. Prediction accuracy for CL, Q, Vc, and Vp using optimal exponents within 1.5-fold of observed values was 78%, 100%, 89%, and 89%, respectively. While optimal exponents for Vc and Vp were similar to conventional exponents, those for CL and Q were lower than that of conventional exponents. Furthermore, no significant difference of prediction accuracy for CL, Q, Vc, and Vp between YTE and LS mutations was observed.
Prediction accuracy of two-compartment model parameters of engineered mAbs in humans using a conventional allometric scaling approach based on cynomolgus monkeys. A CL, B Q, C Vc, D Vp. Solid lines indicate unity. Dashed lines indicate 67% and 150% (± 1.5-fold) of unity. CL clearance, mAbs monoclonal antibodies, Q inter-compartmental clearance, Vc volume of distribution in the central compartment, Vp volume of distribution in the peripheral compartment
Prediction accuracy of two-compartment model parameters of engineered mAbs in humans using an optimized allometric scaling approach based on cynomolgus monkeys. A CL, B Q, C Vc, D Vp. Solid lines indicate unity. Dashed lines indicate 67% and 150% (± 1.5-fold) of unity. CL clearance, mAbs monoclonal antibodies, Q inter-compartmental clearance, Vc volume of distribution in the central compartment, Vp volume of distribution in the peripheral compartment
3.3 Prediction of Plasma mAb Concentration–Time Profiles After IV Injection in Humans
Using an allometric scaling approach with both conventional and optimized exponents, plasma mAb concentration–time profiles for seven engineered mAbs after IV injection in humans were predicted based on those in cynomolgus monkeys. As shown in Figs. 3 and 4, the allometric scaling approach with conventional exponents tended to under-predict plasma mAb concentration in humans. Prediction accuracy within a 2-fold difference of observed values was 68.9% in the allometric scaling approach with conventional exponents. As shown in Fig. 3, plasma concentrations in the elimination phase tended to be especially more under-predicted by the allometric scaling approach with conventional exponents. In contrast, the optimized allometric scaling approach accurately predicted plasma mAb concentrations. It predicted 98.6% of plasma mAb concentrations within a 2-fold difference of observed values. Optimized allometric scaling showed significantly higher prediction accuracy than the conventional approach.
Predicted and observed plasma concentration–time profiles after IV injection in humans using conventional and optimized allometric scaling approaches. Closed squares indicate observed plasma concentration. Dotted lines indicate predicted plasma concentration–time profiles using a conventional allometric scaling approach. Solid lines indicate predicted plasma concentration–time profiles using an optimized allometric scaling approach. A Motavizumab-YTE/MEDI-524-YTE, 0.3 mg/kg. B Motavizumab-YTE/MEDI-524-YTE, 3 mg/kg. C Motavizumab-YTE/MEDI-524-YTE, 15 mg/kg. D Motavizumab-YTE/MEDI-524-YTE, 30 mg/kg. E Amubarvimab/BRII-196, 750 mg. F Amubarvimab/BRII-196, 1500 mg. G Amubarvimab/BRII-196, 3000 mg. H Romlusevimab/BRII-198, 750 mg. I Romlusevimab/BRII-198, 1500 mg. J Romlusevimab/BRII-198, 3000 mg. K Sotrovimab/VIR-7831, 500 mg. L VRC01-LS, 20 mg/kg. M VRC07-523LS, 5 mg/kg. N VRC07-523LS, 20 mg/kg. O VRC07-523LS, 40 mg/kg. P Elipovimab/GS-9722, 150 mg. Q Elipovimab/GS-9722, 500 mg. R Elipovimab/GS-9722, 1000 mg
Relationship of predicted and observed plasma monoclonal antibody (mAb) concentrations. Solid lines indicate unity. A Plasma concentrations in humans predicted using a conventional allometric scaling approach. B Plasma concentrations in humans predicted using an optimized allometric scaling approach
4 Discussion
The effect of mutations to increase the FcRn binding of mAbs in humans was first demonstrated in 2013 [11]. Motavizumab-YTE showed a half-life of around 80 days whereas motavizumab without YTE mutations showed a half-life of only 20–30 days in humans. Based on this impressive report, several engineered mAbs with mutations to increase FcRn binding were evaluated in clinical studies [17,18,19]. In particular, these mutations have been frequently applied to therapeutic anti-virus antibodies [20,21,22]. In fact, the targets of all the engineered mAbs used in this study were viruses, such as respiratory syncytial virus (RSV), human immunodeficiency virus (HIV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This is because long-term efficacy is essential for both the treatment and prophylaxis of virus infection. While several engineered mAbs have been developed and evaluated in clinical studies, a methodology for predicting the pharmacokinetics of engineered mAbs in humans has never been validated. Therefore, in this study, we investigated the optimal exponents for predicting linear two-compartment model parameters (CL, Q, Vc, and Vp) of engineered mAbs in humans from cynomolgus monkeys.
An allometric scaling approach from cynomolgus monkeys has been reported to be valuable for predicting the pharmacokinetics of mAbs in humans [14, 15]. This is because cynomolgus monkey FcRn and human FcRn show similar binding affinity to mAbs with human IgG sequence in the constant region [23]. This similarity of FcRn binding between cynomolgus monkeys and humans has also been reported for engineered mAbs that had significantly stronger FcRn binding affinity than normal mAbs [24]. Therefore, cynomolgus monkey data was used for analysis in this study. As shown in Fig. 5, CL of engineered mAbs between cynomolgus monkeys and humans showed significant positive correlation (p < 0.05, r = 0.81). Previously, a positive correlation of CL for normal mAbs between cynomolgus monkeys and humans has been reported [25, 26]. This significant correlation suggests that CL of engineered mAbs in humans can be predicted from cynomolgus monkeys with the appropriate scaling factor. CL is the most important parameter of the four because CL determines the entire exposure of mAbs. Therefore, prediction accuracy of CL in humans is crucial for predicting pharmacokinetics in humans. In a previous report, the prediction accuracy of CL, Q, Vc, and Vp for normal mAbs using the allometric scaling approach with conventional exponents was 71%, 54%, 88%, and 75% within 1.5-fold of observed values [16]. The observed prediction accuracy for engineered mAbs using the allometric scaling approach with optimized exponents in this study was higher than previous reported for normal mAbs using the allometric scaling approach with conventional exponents. Furthermore, 98.6% of plasma mAb concentrations were accurately predicted within a 2-fold difference of observed values while the allometric scaling approach with conventional exponents achieved only 68.9% prediction accuracy. Additionally, the allometric scaling approach with optimized exponents showed high prediction accuracy across a wide range, from low to high plasma concentrations (from distribution phase to elimination phase). In contrast, the allometric scaling approach with conventional exponents showed a lower prediction accuracy (elimination phase) with low plasma concentrations than with high plasma concentrations (distribution phase). This result suggests that whole plasma mAb concentration–time profiles for engineered mAbs after IV injection in humans can be accurately predicted from cynomolgus monkeys using the optimized allometric scaling approach.
Distribution (A–D) and correlation (E–H) of two-compartment model parameters of engineered mAbs in cynomolgus monkeys and humans. Distribution of CL (A), Q (B), Vc (C), Vp (D) in cynomolgus monkeys (closed triangles) and humans (closed circles). Correlation of CL (E), Q (F), Vc (G), Vp (H) between cynomolgus monkeys and humans. CL clearance, mAbs monoclonal antibodies, Q inter-compartmental clearance, Vc volume of distribution in the central compartment, Vp volume of distribution in the peripheral compartment
This study is the first to recognize that the optimal exponents of allometric scaling for engineered mAbs are different than those for normal mAbs. If allometric scaling with conventional exponents is applied to engineered mAbs to predict human pharmacokinetics, the plasma concentration–time profile in humans will be under-predicted, as shown in Figs. 3 and 4, leading to the inefficient clinical development of engineered mAbs. This is due to over-prediction of CL and Q by the allometric scaling approach with conventional exponents. However, the reason for this difference between normal mAbs and engineered mAbs is still unknown. As described earlier, both normal mAbs and engineered mAbs are reported to show similar FcRn binding affinity in cynomolgus monkeys and humans. Also, endogenous serum IgG concentration has been reported to be around 10–20 mg/mL in both cynomolgus monkeys and humans [27, 28], suggesting that endogenous IgG-mediated FcRn competition would be similar. A mechanistic investigation of this phenomenon needs to be conducted in a future study.
5 Conclusion
This study established an optimized allometric scaling approach for predicting the pharmacokinetics of engineered mAbs in humans. Recently, other than YTE and LS mutations, several mutations to increase FcRn binding have been reported and evaluated in preclinical and clinical studies [29,30,31]. Also, YTE mutations were reported to be applied in ADC [32] and Fc-fusion protein [33] to prolong half-life. The optimized allometric scaling approach could be widely used for mAbs and other therapeutic modalities which contain mutations to increase FcRn binding and can contribute to the efficient development of such therapeutics.
References
Kaplon H, Chenoweth A, Crescioli S, Reichert JM. Antibodies to watch in 2022. MAbs. 2022;14(1):2014296. https://doi.org/10.1080/19420862.2021.2014296.
Labrijn AF, Janmaat ML, Reichert JM, Parren P. Bispecific antibodies: a mechanistic review of the pipeline. Nat Rev Drug Discov. 2019;18(8):585–608. https://doi.org/10.1038/s41573-019-0028-1.
Igawa T, Ishii S, Tachibana T, Maeda A, Higuchi Y, Shimaoka S, et al. Antibody recycling by engineered pH-dependent antigen binding improves the duration of antigen neutralization. Nat Biotechnol. 2010;28(11):1203–7. https://doi.org/10.1038/nbt.1691.
Igawa T, Haraya K, Hattori K. Sweeping antibody as a novel therapeutic antibody modality capable of eliminating soluble antigens from circulation. Immunol Rev. 2016;270(1):132–51. https://doi.org/10.1111/imr.12392.
Mimoto F, Tatsumi K, Shimizu S, Kadono S, Haraya K, Nagayasu M, et al. Exploitation of elevated extracellular ATP to specifically direct antibody to tumor microenvironment. Cell Rep. 2020;33(12): 108542. https://doi.org/10.1016/j.celrep.2020.108542.
Fu Z, Li S, Han S, Shi C, Zhang Y. Antibody drug conjugate: the “biological missile” for targeted cancer therapy. Signal Transduct Target Ther. 2022;7(1):93. https://doi.org/10.1038/s41392-022-00947-7.
Igawa T, Tsunoda H, Tachibana T, Maeda A, Mimoto F, Moriyama C, et al. Reduced elimination of IgG antibodies by engineering the variable region. Protein Eng Des Sel. 2010;23(5):385–92. https://doi.org/10.1093/protein/gzq009.
Bumbaca Yadav D, Sharma VK, Boswell CA, Hotzel I, Tesar D, Shang Y, et al. Evaluating the use of antibody variable region (Fv) charge as a risk assessment tool for predicting typical cynomolgus monkey pharmacokinetics. J Biol Chem. 2015;290(50):29732–41. https://doi.org/10.1074/jbc.M115.692434.
Dall’Acqua WF, Kiener PA, Wu H. Properties of human IgG1s engineered for enhanced binding to the neonatal Fc receptor (FcRn). J Biol Chem. 2006;281(33):23514–24. https://doi.org/10.1074/jbc.M604292200.
Zalevsky J, Chamberlain AK, Horton HM, Karki S, Leung IW, Sproule TJ, et al. Enhanced antibody half-life improves in vivo activity. Nat Biotechnol. 2010;28(2):157–9. https://doi.org/10.1038/nbt.1601.
Robbie GJ, Criste R, Dall’acqua WF, Jensen K, Patel NK, Losonsky GA, et al. A novel investigational Fc-modified humanized monoclonal antibody, motavizumab-YTE, has an extended half-life in healthy adults. Antimicrob Agents Chemother. 2013;57(12):6147–53. https://doi.org/10.1128/AAC.01285-13.
Loo YM, McTamney PM, Arends RH, Abram ME, Aksyuk AA, Diallo S, et al. The SARS-CoV-2 monoclonal antibody combination, AZD7442, is protective in nonhuman primates and has an extended half-life in humans. Sci Transl Med. 2022;14(635):eabl8124. https://doi.org/10.1126/scitranslmed.abl8124.
Gaudinski MR, Houser KV, Doria-Rose NA, Chen GL, Rothwell RSS, Berkowitz N, et al. Safety and pharmacokinetics of broadly neutralising human monoclonal antibody VRC07-523LS in healthy adults: a phase 1 dose-escalation clinical trial. Lancet HIV. 2019;6(10):e667–79. https://doi.org/10.1016/S2352-3018(19)30181-X.
Deng R, Iyer S, Theil FP, Mortensen DL, Fielder PJ, Prabhu S. Projecting human pharmacokinetics of therapeutic antibodies from nonclinical data: what have we learned? MAbs. 2011;3(1):61–6. https://doi.org/10.4161/mabs.3.1.13799.
Dong JQ, Salinger DH, Endres CJ, Gibbs JP, Hsu CP, Stouch BJ, et al. Quantitative prediction of human pharmacokinetics for monoclonal antibodies: retrospective analysis of monkey as a single species for first-in-human prediction. Clin Pharmacokinet. 2011;50(2):131–42. https://doi.org/10.2165/11537430-000000000-00000.
Haraya K, Tachibana T, Nezu J. Quantitative prediction of therapeutic antibody pharmacokinetics after intravenous and subcutaneous injection in human. Drug Metab Pharmacokinet. 2017;32(4):208–17. https://doi.org/10.1016/j.dmpk.2017.05.002.
Yu XQ, Robbie GJ, Wu Y, Esser MT, Jensen K, Schwartz HI, et al. Safety, tolerability, and pharmacokinetics of MEDI4893, an investigational, extended-half-life, anti-Staphylococcus aureus alpha-toxin human monoclonal antibody, in healthy adults. Antimicrob Agents Chemother. 2017. https://doi.org/10.1128/AAC.01020-16.
Gaudinski MR, Berkowitz NM, Idris AH, Coates EE, Holman LA, Mendoza F, et al. A monoclonal antibody for malaria prevention. N Engl J Med. 2021;385(9):803–14. https://doi.org/10.1056/NEJMoa2034031.
Hussaini A, Mukherjee R, Berdieva DM, Glogowski C, Mountfield R, Ho PTC. A double-blind, phase I, single ascending dose study to assess the safety, pharmacokinetics, and pharmacodynamics of BOS161721 in healthy subjects. Clin Transl Sci. 2020;13(2):337–44. https://doi.org/10.1111/cts.12715.
Lanini S, Milleri S, Andreano E, Nosari S, Paciello I, Piccini G, et al. Safety and serum distribution of anti-SARS-CoV-2 monoclonal antibody MAD0004J08 after intramuscular injection. Nat Commun. 2022;13(1):2263. https://doi.org/10.1038/s41467-022-29909-x.
Griffin MP, Khan AA, Esser MT, Jensen K, Takas T, Kankam MK, et al. Safety, tolerability, and pharmacokinetics of MEDI8897, the respiratory syncytial virus prefusion F-targeting monoclonal antibody with an extended half-life, in healthy adults. Antimicrob Agents Chemother. 2017. https://doi.org/10.1128/AAC.01714-16.
Aliprantis AO, Wolford D, Caro L, Maas BM, Ma H, Montgomery DL, et al. A phase 1 randomized, double-blind, placebo-controlled trial to assess the safety, tolerability, and pharmacokinetics of a respiratory syncytial virus neutralizing monoclonal antibody MK-1654 in healthy adults. Clin Pharmacol Drug Dev. 2021;10(5):556–66. https://doi.org/10.1002/cpdd.883.
Abdiche YN, Yeung YA, Chaparro-Riggers J, Barman I, Strop P, Chin SM, et al. The neonatal Fc receptor (FcRn) binds independently to both sites of the IgG homodimer with identical affinity. MAbs. 2015;7(2):331–43. https://doi.org/10.1080/19420862.2015.1008353.
Borrok MJ, Wu Y, Beyaz N, Yu XQ, Oganesyan V, Dall’Acqua WF, et al. pH-dependent binding engineering reveals an FcRn affinity threshold that governs IgG recycling. J Biol Chem. 2015;290(7):4282–90. https://doi.org/10.1074/jbc.M114.603712.
Tam SH, McCarthy SG, Brosnan K, Goldberg KM, Scallon BJ. Correlations between pharmacokinetics of IgG antibodies in primates vs. FcRn-transgenic mice reveal a rodent model with predictive capabilities. MAbs. 2013;5(3):397–405. https://doi.org/10.4161/mabs.23836.
Hotzel I, Theil FP, Bernstein LJ, Prabhu S, Deng R, Quintana L, et al. A strategy for risk mitigation of antibodies with fast clearance. MAbs. 2012;4(6):753–60. https://doi.org/10.4161/mabs.22189.
Nixon AE, Chen J, Sexton DJ, Muruganandam A, Bitonti AJ, Dumont J, et al. Fully human monoclonal antibody inhibitors of the neonatal fc receptor reduce circulating IgG in non-human primates. Front Immunol. 2015;6:176. https://doi.org/10.3389/fimmu.2015.00176.
Kiessling P, Lledo-Garcia R, Watanabe S, Langdon G, Tran D, Bari M, et al. The FcRn inhibitor rozanolixizumab reduces human serum IgG concentration: a randomized phase 1 study. Sci Transl Med. 2017. https://doi.org/10.1126/scitranslmed.aan1208.
Paguntalan H, Magyarics Z, Connolly LE, Hershberger E, Narayan K, Gupta D, et al. 633. Preliminary results from a phase 1 single ascending-dose study assessing safety, serum viral neutralizing antibody titers (sVNA), and pharmacokinetic (PK) profile of ADG20: an extended half-life monoclonal antibody being developed for the treatment and prevention of coronavirus disease (COVID-19). Open Forum Infect Dis. 2021;8(Supplement_1):S420-S. https://doi.org/10.1093/ofid/ofab466.831.
Jacobson PB, Goody R, Lawrence M, Mueller BK, Zhang X, Hooker BA, et al. Elezanumab, a human anti-RGMa monoclonal antibody, promotes neuroprotection, neuroplasticity, and neurorecovery following a thoracic hemicompression spinal cord injury in non-human primates. Neurobiol Dis. 2021;155: 105385. https://doi.org/10.1016/j.nbd.2021.105385.
Maeda A, Iwayanagi Y, Haraya K, Tachibana T, Nakamura G, Nambu T, et al. Identification of human IgG1 variant with enhanced FcRn binding and without increased binding to rheumatoid factor autoantibody. MAbs. 2017;9(5):844–53. https://doi.org/10.1080/19420862.2017.1314873.
Scribner JA, Hicks SW, Sinkevicius KW, Yoder NC, Diedrich G, Brown JG, et al. Preclinical evaluation of IMGC936, a next generation maytansinoid-based antibody-drug conjugate targeting ADAM9-expressing tumors. Mol Cancer Ther. 2022. https://doi.org/10.1158/1535-7163.Mct-21-0915.
Douthwaite J, Moisan J, Privezentzev C, Soskic B, Sabbah S, Cohen S, et al. A CD80-biased CTLA4-Ig fusion protein with superior in vivo efficacy by simultaneous engineering of affinity, selectivity, stability, and FcRn binding. J Immunol. 2017;198(1):528–37. https://doi.org/10.4049/jimmunol.1600682.
The Therapeutic Goods Administration in the Australian Government Department of Health. Tixagevimab/cilgavimab (Evusheld). 2022. https://www.tga.gov.au/sites/default/files/auspar-tixagevimab-cilgavimab-220311.pdf. Accessed 1 June 2022.
Patent. WO2021185346. ANTI-SARS-COV-2 ANTIBODIES AND USES THEREOF.
Zhang Y, Hao X, Ma J, Wang M, Li Y, Liu Y, et al. Phase 1 safety and pharmacokinetics studies of BRII-196 and BRII-198, SARS-CoV-2 spike-targeting monoclonal antibodies. medRxiv. 2021:2021.07.21.21260964. https://doi.org/10.1101/2021.07.21.21260964.
Pharmaceutical and Medical Devices Agency (PMDA). Sotrovimab (Xevudy). 2021. https://www.pmda.go.jp/drugs/2021/P20210927001/index.html. Accessed 1 June 2022.
Ko SY, Pegu A, Rudicell RS, Yang ZY, Joyce MG, Chen X, et al. Enhanced neonatal Fc receptor function improves protection against primate SHIV infection. Nature. 2014;514(7524):642–5. https://doi.org/10.1038/nature13612.
Yang J, Cunningham CK, McFarland EJ, Mascola JR, Graham BS, Ledgerwood JE, et al. Population pharmacokinetics of vrc01ls in term infants and adults. CROI-2021.
HPTN. A multicenter, randomized, partially blinded phase 1 clinical trial to evaluate the safety and serum concentrations of a human monoclonal antibody, VRC-HIVMAB075-00-AB (VRC07-523LS), administered in multiple doses and routes to healthy, HIV-uninfected adults. 2018. https://www.hptn.org/research/studies/127_087. Accessed 1 June 2022.
Patent. WO2017106346. Human immunodeficiency virus neutralizing antibodies.
Ruane P, Daar E, Workowski K, Begley R, Humeniuk R, Makadzange T et al. Safety & pharmacokinetics of GS-9722 in HIV-negative participants and people with HIV. CROI-2020.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This work was fully supported by Chugai Pharmaceutical Co. Ltd.
Conflict of interest
Kenta Haraya and Tatsuhiko Tachibana are employees of Chugai Pharmaceutical Co. Ltd. and declare that they have no potential conflicts of interest that might be relevant to the contents of this manuscript.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
Not applicable.
Code availability
Not applicable.
Author’s contributions
Kenta Haraya and Tatsuhiko Tachibana conducted data collection and analysis and wrote the manuscript.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Haraya, K., Tachibana, T. Translational Approach for Predicting Human Pharmacokinetics of Engineered Therapeutic Monoclonal Antibodies with Increased FcRn-Binding Mutations. BioDrugs 37, 99–108 (2023). https://doi.org/10.1007/s40259-022-00566-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40259-022-00566-2