Abstract
Persian shallot, Allium hirtifolium Boiss. (AH), is an Iranian native medicinal plant belongs to Alliaceae family. Here, we investigated in vitro antibacterial activity of hydro-alcoholic extract derived from bulbs of AH. We also employed in silico molecular docking to decipher mechanisms of its antibacterial effects. Minimum inhibitory concentrations (MIC) and minimum bactericidal concentration (MBC) against E. coli ATCC 25922 were determined. Molecular docking was performed for major phytochemicals of AH against ribosome recycling factor (RRF). E. coli ATCC 25922 was gentamicin-resistant while AH showed MIC (42 ± 18 μg/ml) and MBC (106 ± 36 μg/ml) against E. coli. In silico results reported all phytochemicals of AH shown acceptable negative binding affinity (kcal/mol) with RRF. In essence, the binding affinities of alliogenin (−11.6), gitogenin (−11.6), kaempferol (−10.2), linoleic acid (−8.4), oleic acid (−8.0), palmitic acid (−7.4), palmitoleic acid (−8.4), quercetin (−10.8), and shallomin (−13.4) with RRF were comparable to that of gentamicin (−12.6). In sum, hydro-alcoholic extract of bulbs of AH could be considered as a commercial phytobiotics if in-depth antibacterial assays employed in future studies. More interestingly, shallomin showed more promising binding affinity with RRF and can be considered as lead molecule for future drug discovery.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Secondary metabolites, phytochemicals, are bioactive compounds derived from medicinal plants and able to destroy pathogenic bacteria via various toolboxes (Monte et al. 2014). In this line, antibacterial phytochemicals, phytobiotics, or synthetic antibiotics block or kill pathogenic microorganisms by targeting proteins, genes, and metabolites that are necessary for survival of microorganisms. For instance, ribosome recycling factor (RRF) is a protein present in E. coli responsible for separation of ribosomes from mRNA after the termination of translation and reuse them (Janosi et al. 1994). Indeed, gentamicin is a classic antibiotic that irreversibly binds to 30S subunit of bacterial ribosome and abolishes protein synthesis, however we encounter to the emergence of gentamicin-resistance bacteria for decades (Weinstein 1967). Therefore, the seeking phytobiotics that work on multi-drug resistant bacteria would be a necessity and economic option nowadays.
Allium hirtifolium Boiss. (AH), commonly known as Persian shallot, is a medicinal plant belongs to family Alliaceae and native to Iran which overtly used for the treatment of many diseases (Jafariana et al. 2003). Phytochemical compounds which are reported in the bulbs of AH including linolenic, linoleic, palmitoleic, palmitic, oleic and stearic acid, kaempferol, quercetin, shallomin, furostanal, spirostanal, sulphur containing compounds (thiosulfinates), and flavonoids (Jafariana et al. 2003; Amin and Kapadnis 2005; Barile et al. 2005; Ebrahimi et al. 2008; Kazemi et al. 2010; Amin et al. 2012; Asgarpanah and Ghanizadeh 2012). In this continuum, aqueous extracts of bulbs of AH showed considerable inhibitory effect against pathogenic bacteria including Staphylococcus aureus, Enterococcus faecalis, Bacillus subtilis, E. coli, Pseudomonas aeruginosa, and Klebsiella pneumonia (Ghahremani-majd et al. 2012).
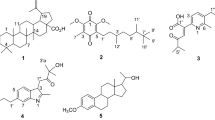
Based on (Kurdish) ethnomedicine, this study was aimed firstly to probe in vitro antibacterial activity of the bulbs of AH against E. coli and secondly to decipher in silico docking of its major reported phytochemicals (Fig. 1) against RRF as a target.
Methods
Plant preparation
The Persian shallot collected from Iran and Iraq since March to October 2016. The collected specie was authenticated by botanist, third author, as Allium hirtifolium Boiss (AH). To prepare hydro-alcoholic extract, powdered bulbs (10 g) were extracted twice with 100 ml of 70% ethanol for 48 h at room temperature. The extracted suspensions were filtered and resulting filtrates were dried using a rotary evaporator and stored at −20 °C until further use. For the antibacterial assays, dried extract was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 100 mg/ml and stored at 4 °C as stock solutions.
In vitro antibacterial assay
Gentamicin was used to assay the effects of antibiotics on the strains E. coli (ATCC® 25922TM) at 5 µg/ml for each well of microtiter plate. E. coli was cultured on Mueller–Hinton Agar (MHA) and Mueller–Hinton Broth (MHB) media. The antibacterial activity of bulbs of AH on E. coli was determined as minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) using broth microdilution method (Wiegand et al. 2008). Serial dilutions of the extract were prepared in a 96-well microtiter plate. To each well, 100 μl of indicator solution that prepared by dissolving a 10 mg extract in 1 ml of DMSO and 100 μl of MHB were added. Finally, 50 μl of bacterial suspension (106 colony forming unite (CFU)/ml) were added to each well to achieve a concentration of 104 CFU/ml. The microtiter plates were prepared in triplicates and incubated at 37 °C for 18–24 h.
In silico antibacterial assay
Molecular docking of selected bioactive compounds of AH (Fig. 1) and RRF target protein was performed by docking program (VINA WIZARD) installed on PyRx software ver. 0.8 (Dallakyan and Olson 2015). For docking studies, crystal structure of RRF was taken from the Protein Data Bank (PDB) at the Research Collaboratory for Structural Bioinformatics (http://www.RCSB.org). The PDB format of target protein has been prepared for docking using Molegro Virtual Docker (Thomsen and Christensen 2006) and Chimera 1.8.1 (http://www.rbvi.ucsf.edu/chimera) softwares before submission to PyRx software. The structures of the major phytocompounds of AH were retrieved from ZINC database ver. 12.0 (http://zinc.docking.org/) and DrugBank ver. 5.0 (https://www.drugbank.ca/) or drawn by ChemDraw (http://www.cambridgesoft.com/software/overview.aspx) as required.
The docking results were shown as binding affinity (kcal/mol) values. More negative the binding affinity means better docking of ligand in binding site. The best fitted conformer of ligand has been combined with target protein in Molegro Virtual Docker (Thomsen and Christensen 2006) or Chimera 1.8.1 (http://www.rbvi.ucsf.edu/chimera) and its atomic interaction has been analysed with LigPlot+ software (Laskowski and Swindells 2011). In LigPlot+, purple lines show ligand bonds; orange lines show non-ligand bonds; dotted lines show hydrogen bonds and its length; half red circle show non-ligand residues involved in hydrophobic contacts; and black dots show corresponding atoms involved in hydrophobic contacts (vide infra).
Results and discussion
In numerous developing countries, traditional medicine is still the mainstay of therapeutics. Even in developed countries, raw materials for industrialization of base drugs are extracted from medicinal plants. In this regard, (reverse) pharmacognosy has played a pivotal role in the detection and development of novel drugs and therapies (Satyajit 2012). Medicinal plants are rich in a wide array of natural components which have antimicrobial properties and could be utilized as a valuable source for phytobiotics (Cowan 1999). Meanwhile, E. coli causes diverse diseases in digestive tract in human and animals (Kaper et al. 2004). Strain of E. coli ATCC 25922 employed in this study was resistant to gentamicin. Gentamicin is a bactericidal and massively used antibiotic that causes mRNA decoding mistake, prevents transfer RNA and mRNA translocation, and blocks ribosome recycling in bacteria (Borovinskaya et al. 2007). Our results showed that this strain of E. coli ATCC 25922 was resistant to gentamicin while sensitive to AH. In essence, bulbs of AH have inhibitory at MIC 42 ± 18 (μg/ml) and bactericidal effects at MBC 106 ± 36 (μg/ml) on E. coli. With respect to antimicrobial activity, the extracts of bulbs AH showed moderate to good inhibitory activities against pathogenic bacteria including Bacillus cereus, Listeria monocytogenes, Proteus vulgaris, and Salmonella typhimurium (Ghasemi et al. 2016).
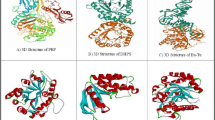
In this study, we selected gentamicin as reference antibiotic to compare its in silico binding affinity with RRF (PDB:1WIH) to those of ligands of bulbs of AH. Here we decided to decipher computationally phytocompounds which may involve in anti-E coli effects of AH (Table 1, Fig. 2). More specifically, we selected RRF as target protein since it is product of the frr gene in E. coli and is responsible for separation of ribosomes from mRNA after the termination of translation and recycling of ribosomes (Janosi et al. 1994).
The molecular docking of phytochemicals of Allium hirtifolium Boiss (in yellow) against ribosome recycling factor (PDB:1WIH; in cyan). Alliogenin Corresponding atoms and non-ligand residues IIe74, Asn60, Pro75, Pro33, Pro73, Lys76, Gln34, Val35, and Lue61 involved in hydrophobic interactions. Hydrogen bonds between alliogenin and Arg71 shown by dotted red lines. Gitogenin Corresponding atoms and non-ligand residues IIe74, Pro33, Pro73, Lys76, Gln34, and Val35 involved in hydrophobic interactions. Hydrogen bonds between gitogenin and Arg71 and Glu64 shown by dotted red lines. Kaempferol Hydrogen bonds between kaempferol and Thr12, His10, and Gly58 shown by dotted red lines. Linoleic acid Hydrogen bonds between linoleic acid and Ala41 and Ser42 shown by dotted red line. Oleic acid Hydrogen bonds between oleic acid and Ala41 shown by dotted red line. Palmitic acid Hydrogen bonds between palmitic acid and Ala16 shown by dotted red line. Palmitoleic acid Hydrogen bonds between palmitoleic acid and Ala41 shown by dotted red line. Quercetin Corresponding atoms and non-ligand residues Ala41, Met40, Gly26, Gln27, and Asn23 involved in hydrophobic interactions. Hydrogen bonds between quercetin and Gln24, IIe25, Asn39 and Ser42 shown by dotted red lines. Shallomin Corresponding atoms and non-ligand residues Thr12 and IIe11 involved in hydrophobic interactions. Hydrogen bonds between shallomin and Glu56, Ala53, and Ser57 shown by dotted red lines. Gentamicin Corresponding atoms and non-ligand residues IIe11 involved in hydrophobic interactions. Hydrogen bonds between gentamicin and Ser57, Thr12 and His10 shown by dotted red lines
Based on docking results, gentamicin showed negative binding affinity with RRF at −12.6 kcal/mol (Table 1) and docked by hydrophobic interactions and hydrogen bonds (Fig. 2). In silico findings showed that among phytocompounds found in bulbs of AH, shallomin has been docked with lower binding affinity with RRF with respect to gentamicin. Shallomin is a flavonoid and would be a new lead molecule in AH and has a wide range of antibacterial properties (Bouktaib et al. 2002). Shallomin showed the lowest negative binding affinity against RRF among phytocompounds of AH. It docked with RRF utilizing both hydrophobic interactions and hydrogen bonds. Shallomin has been docked with amino acid residues (Thr12 and Ser57) in RRF which gentamicin also employed these amino acids to dock with RRF (Fig. 2). In a recent study, a preliminary in vivo toxicological estimate of escalating doses of shallomin was made and showed that shallomin was a relatively safe agent when administrated at its standard antimicrobial concentration via intraperitoneal route (Amin et al. 2012).
Quercetin is a major representative of the flavonol subclass and often found as its glycoside form which has antibacterial activity (Kumar et al. 2016). Quercetin showed in vitro antibacterial activity against Bacillus subtilis, E. coli, Staphylococcus aureus, and Salmonella typhimurium as major food-borne microorganisms (Mertens-Talcott and Percival 2005). In the present study, quercetin has been suitably docked with RRF via both hydrophobic interaction and hydrogen bonds. The hydroxyl groups of quercetin participated in four hydrogen bonds with various amino acids of RRF (Fig. 2).
Kaempferol is a type of flavonoid found in various plants and plant-derived formulations and used in traditional medicine. Numerous preclinical researches have shown that kaempferol has antimicrobial, antioxidant, anticancer, anti-inflammatory, antidiabetic, cardioprotective, neuroprotective, anti-osteoporotic, anxiolytic, analgesic, and anti-allergic activities (Calderón-Montaño et al. 2011). In this study, kaempferol like its congener, quercetin, has been docked with RRF by using four hydrogen bonds (Fig. 2). More surprisingly, kaempferol like gentamicin has been docked with identical amino acids (Thr12 and His10) of RRF. In the other word, kaempferol mimics the strategy of gentamicin to dock RRF (Fig. 2).
Oleic acid is a monounsaturated ω-9 fatty acid widely distributed in nature (De Silva et al. 2014). Oleic acid is a bactericidal agent against important pathogenic microorganisms including methicillin-resistant Staphylococcus aureus and Helicobacter pylori (Farrington et al. 1992). Oleic acid has been moderately docked with Ala41 residue of RRF as compared to other fatty acids of AH (Fig. 2).
Linoleic acid is a polyunsaturated omega-6 fatty acid occurring widely in plant glycosides (Pariza et al. 2000). Linoleic acid has antibacterial activities against Bacillus megaterium, Pseudomonas phaseolicola and Streptococcus mutans (Dilika et al. 2000). Linoleic acid has been bound with amino groups of Ala41 and Ser42 residues of RRF through its carboxyl group. Its binding affinity was more suitable as compared with other fatty acids reported in this study.
Palmitic acid is the most prevalent saturated fatty acid present in plants, animals and microorganisms (Gunstone et al. Gunstone et al. 2007). This fatty acid destroys the structure and function of bacterial cell walls and membranes (McGaw et al. 2002). Palmitic acid showed the weakest binding affinity with RRF among fatty acids and all ligands of AH in this study (Table 1). It also has been docked through its carboxyl group with amino group of Ala16 in RRF (Fig. 2).
Palmitoleic acid is a class of unsaturated fatty acid synthesized from the breakdown of triglyceride. Palmitoleic acid has bactericidal properties toward Staphylococcus aureus, Helicobacter pylori, Streptococcus salivarius, Fusobacterium nucleatum and Neisseria gonorrhoeae (Yamamoto et al. 2015). Palmitoleic acid and linoleic acid showed identical binding affinity to RRF (Table 1) although palmitoleic acid has been solely docked with Ala41 of RRF (Fig. 2). In sum, all carboxylic groups of fatty acids reported in AH have been docked via hydrogen bonds to amino group of amino acid Ala or Ser of RRF (Fig. 2). To the best of our knowledge, this study for the first time shows that interaction of fatty acids and RRF. Further investigations are welcomed to decipher the role of Ala41, Ala16, and Ser42 in RRF that interact with carboxyl groups of fatty acids.
Alliogenin and gitogenin are type of spirostanol saponins (Amin and Kapadnis 2005). Alliogenin and gitogenin shows identical and very powerful binding affinity against RRF (Table 1). Based on the similar amino acids of RRF which participated in hydrophobic interactions and hydrogen bond formation with alliogenin and gitogenin, we can conclude that these two ligands would be docked in identical site of RRF. Alliogenin also has been isolated from the bulbs of Allium minutiflorum and has antifungal activity (Barile et al. 2007). These spirostanol saponins make hydrogen bonds between hydroxyl groups of their spirostan rings and guanidinium group of Arg71 of RRF. Gitogenin additionally formed hydrogen bond with delta carboxyl group of Glu64 residue of RRF. The antimicrobial activity of gitogenin has been not reported previously, while our in silico results show that this bioactive compound would be a lead molecule in antibacterial investigations.
Conclusions
The emergence of antibiotic-resistant bacteria and hard access to antibiotics in distinct parts of the world motivated researchers to seek alternate therapeutics. In this continuum, hydro-alcoholic extract prepared of AH bulbs showed promising antibacterial activity against E. coli. In silico findings showed that among phytochemical compounds found in bulbs of AH, shallomin has more negative binding affinity to RRF compared to gentamicin and would be considered as a lead molecule for further attempts to design phytobiotics.
References
Amin M, Kapadnis BP (2005) Heat stable antimicrobial activity of Allium ascalonicum against bacteria and fungi. Indian J Exp Biol 43:751–754
Amin A, Pipelzadeh MH, Mehdinejad M, Rashidi I (2012) An in vivo toxicological study upon shallomin, the active antimicrobial constitute of Persian shallot (Allium hirtifolium, Boiss) extract. Jundishapur J Nat Pharm Prod 7:17–21
Asgarpanah J, Ghanizadeh B (2012) Pharmacologic and medicinal properties of Allium hirtifolium Boiss. Afr J Pharm Pharmacol 6:1809–1814
Barile E, Capasso R, Izzo AA, Lanzotti V, Sajjadi E, Zolfaghari B (2005) Structure-activity relationships for saponins from Allium hirtifolium and Allium elburzense and their antispasmodic activity. Planta Med J 71:1010–1018
Barile E, Bonanomi G, Antignani V, Zolfaghari B, Sajjadi SE, Scala F, Lanzotti V (2007) Saponins from Allium minutiflorum with antifungal activity. Phytochemistry 68(5):596–603
Borovinskaya AM, Pai DR, Zhang W, Schuwirth SB, Holton MJ, Hirokawa G, Kaji H, Kaji A, Cate JH (2007) Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat Struct Mol Biol J 14:727–732
Bouktaib M, Aziz A, Christian R (2002) Regio- and stereo-selective synthesis of the major metabolite of quercetin, quercetin-3-o-b-d-glucuronide. Tetrahedron Lett J. 43:6263–6266
Calderón-Montaño JM, Burgos-Morón E, Pérez-Guerrero C, López-Lázaro M (2011) A review on the dietary flavonoid kaempferol. Mini Rev Med Chem J 11:298–344
Cowan MM (1999) Plant products as antimicrobial agents. Clin Microbiol Rev 12:564–582
Dallakyan S, Olson AJ (2015) Small-molecule library screening by docking with PyRx. Methods Mol Biol J 63:243–450
de Silva PS, Luben R, Shrestha SS, Khaw KT, Hart AR (2014) Dietary arachidonic and oleic acid intake in ulcerative colitis etiology: a prospective cohort study using 7-day food diaries. Eur J Gastroenterol Hepatol 1:11–18
Dilika FK, Bremner PD, Meyer JJM (2000) A synergistic effect between the two fatty acids was observed against Staphylococcus aureus and Micrococcus. Fitoterapia J 1:450–452
Ebrahimi R, Zamani ZA, Kashi AAK, Jabari A (2008) Comparison of fatty acid and minerals content of seventeen Allium hirtifolium Boiss. Iran Food Sci Tech J 56:61–68
Farrington M, Brenwald N, Haines D, Walpole E (1992) Resistance to desiccation and skin fatty acids in outbreak strains of methicillin-resistant Staphylococcus aureus. J Med Microbiol 36:56–60
Ghahremani-majd H, Dashti F, Dastan D, Mumivand H, Hadian J, Esna-Ashari M (2012) Antioxidant and antimicrobial activities of Iranian mooseer (Allium hirtifolium Boiss) population. Hortic Environ Biotech J 2:116–122
Ghasemi PA, Ahmadzadeh Y, Malekpoor F (2016) Variation in antioxidant, and antibacterial activities and total phenolic content of the bulbs of mooseer (Allium hirtifolium Boiss). Acta Agric Slov 1:34–41
Gunstone FD, Harwood JL, Dijkstra AJ (2007) The lipid handbook with CD-ROM, vol 22, 3rd edn. CRC Press, Boca Raton, pp 1–800
Jafariana A, Ghannadib A, Elyasia A (2003) The effects of Allium hirtifolium Boiss. on cell-mediated immune response in mice. Iran. J Pharm Res 2:51–55
Janosi L, Shimizu I, Kaji A (1994) Ribosome recycling factor (ribosome releasing factor) is essential for bacterial growth. Proc Nat Acad Sci USA 91:53–4249
Kaper BJ, Nataro PJ, Mobley LH (2004) Pathogenic Escherichia coli. Nat Rev Microbiol 2:124–140
Kazemi S, Asgary S, Moshtaghian J, Rafieian M, Adelnia A, Shamsi F (2010) Liver protective effects of hydroalcoholic extract of Allium hirtifolium Boiss. In rats with alloxan-induced diabetes mellitus. Arya Atheroscler 6:11–15
Kumar VD, Verma PRP, Singh SK (2016) Morphological and in vitro antibacterial efficacy of quercetin loaded nanoparticles against food-borne microorganisms. LWT Food Sci Technol 66:638–650
Laskowski RA, Swindells MB (2011) LigPlot+: Multiple ligand–protein interaction diagrams for drug discovery. J Chem Inf Model 51:2778–2786
McGaw LJ, Jäger AK, Van SJ (2002) Antibacterial effects of fatty acids and related compounds from plants. S Afr J Bot 68:417–423
Mertens-Talcott SU, Percival SS (2005) Ellagic acid and quercetin interact synergistically with resveratrol in the induction of apoptosis and cause transient cell cycle arrest in human leukemia cells. Cancer Lett J 218:141–151
Monte J, Abreu AC, Borges A, Simões LC, Simões M (2014) Antimicrobial activity of selected phytochemicals against Escherichia coli and Staphylococcus aureus and their biofilms. Pathogens 3:473–498
Pariza MW, Park Y, Cook ME (2000) Mechanisms of action of conjugated linoleic acid: evidence and speculation. Proc Soc Exp Biol Med J 223:8–13
Satyajit DS (2012) Pharmacognosy in modern pharmacy curricula. Pharmacogn Mag J 8:91–92
Thomsen R, Christensen MH (2006) Mol Dock: a new technique for high–accuracy molecular docking. J Med Chem 49:3315–3321
Weinstein MJ (1967) Biological activity of the antibiotic components of the gentamicin complex. J Bacteriol 3:789–790
Wiegand I, Hilpert K, Hancock RE (2008) Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc J 3:163–175
Yamamoto Y, Kawamura Y, Yamazaki Y, Kijima T, Morikawa T, Nonomura Y (2015) Palmitoleic acid calcium salt: a lubricant and bactericidal powder from natural lipids. J Oleo Sci 64:8–283
Acknowledgments
This paper emanates from MSc thesis of first author submitted to Department of Biology, Faculty of Science, Razi University 67149-67346, Kermanshah, Iran. Authors acknowledge personnel of Laboratory of Microbiology for technical assistance. This study was supported by intramural fund and first and fourth authors paid the fee of in silico investigation.
Authors contributions statement
NK gathered data of Kurdish ethnomedicine and authenticated plants. SEA, KC, IK, and NK carried out the experiments. SEA and IK analyzed data and carried out the molecular docking work. SEA and IK prepared the manuscript while all authors have read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ali, S.E., Chehri, K., Karimi, N. et al. Computational approaches to the in vitro antibacterial activity of Allium hirtifolium Boiss against gentamicin-resistant Escherichia coli: focus on ribosome recycling factor. In Silico Pharmacol. 5, 7 (2017). https://doi.org/10.1007/s40203-017-0027-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s40203-017-0027-z