Abstract
Future machine learning strategies for materials process optimization will likely replace human capital-intensive artisan research with autonomous and/or accelerated approaches. Such automation enables accelerated multimodal characterization that simultaneously minimizes human errors, lowers costs, enhances statistical sampling, and allows scientists to allocate their time to critical thinking instead of repetitive manual tasks. Previous acceleration efforts to synthesize and evaluate materials have often employed elaborate robotic self-driving laboratories or used specialized strategies that are difficult to generalize. Herein we describe an implemented workflow for accelerating the multimodal characterization of a combinatorial set of 915 electroplated Ni and Ni–Fe thin films resulting in a data cube with over 160,000 individual data files. Our acceleration strategies do not require manufacturing-scale resources and are thus amenable to typical materials research facilities in academic, government, or commercial laboratories. The workflow demonstrated the acceleration of six characterization modalities: optical microscopy, laser profilometry, X-ray diffraction, X-ray fluorescence, nanoindentation, and tribological (friction and wear) testing, each with speedup factors ranging from 13–46x. In addition, automated data upload to a repository using FAIR data principles was accelerated by 64x.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Materials science has historically progressed via artisan-style research relying on scientists to synthesize and characterize samples individually at a large human capital cost. Accelerating materials synthesis and characterization would enable wider sampling of fabrication variable spaces and statistical treatment of data enabled by multiple replicates, machine learning, multi-variate regression, support vector machines, and other methods.
New frontiers in materials discovery are being explored with machine learning and have already aided discovery in areas like ultraincompressible superhard materials [1], binary alloy crystallography [2], and polyelemental heterostructures [3]. Large, consistent datasets can provide machine learning methods with the required volume of data for training, testing, and validation. Existing training datasets include materials databases populated with experimentally and computationally derived data [4,5,6,7,8,9]. However, the curation of experimental, multimodal data cubes characterizing a population of samples is still rather rare, and few authors make their data available publicly [10].
Investment in task automation is one strategy which could accelerate multimodal characterization and minimize human errors, lower costs, and allow scientists to allocate their time to critical thinking as opposed to repetitive manual tasks. Few commercially available instruments employ comprehensive automation functionalities.
To fill this gap, current research has focused on creating custom solutions for key fronts, including materials synthesis [11,12,13,14,15], materials characterization [12, 16,17,18], and materials simulations [1, 19,20,21,22,23,24,25]. Despite recent advances in this emerging field, a vast majority of materials research is still conducted in piecewise artisan modalities limited by human expert-based instrument operators.
Herein we describe an implemented workflow for accelerating the characterization of thin films. Our acceleration strategies do not require manufacturing-scale resources (e.g., extensive robotic workcells, etc.) and are thus amenable to typical materials research facilities in academic, government, or commercial laboratories. To avoid building custom instrumentation, the workflow leverages existing instrumentation features and addresses their weaknesses with time-saving strategies that can be implemented in whole or part by others, dependent on project needs and budget. In contrast, others have employed costly self-driving laboratories [26, 27] and used specialized strategies that are difficult to generalize [17, 18, 17,28,29,30,31,32,33,34,35,18]. For instance, high-throughput XRD has been reported for samples specifically co-deposited on wafers [33, 35], which is limited to only specific processes. There are numerous reports of custom-built high-throughput characterization systems such as a high-throughput UV–Vis spectrometer [34], optical scanning droplet cells to collect photoelectrochemical data [17, 18, 36], scanning electrochemical cells [28, 30, 31], a millifluidic system for electrochemical corrosion testing [32], robotics for sample handling [26, 27, 37], and flow reactors for synthesizing and characterizing quantum dots [29]. While these tools elegantly solve workflow challenges, the solutions presented would be difficult to implement on pre-existing instrumentation.
The workflow described herein leverages multimodal characterization methods to analyze the crystal structure, composition, visible appearance, and surface morphology of metallic films—resulting in a set of material properties that can be traced back to their corresponding processing conditions. A similar accelerated multimodal workflow was developed previously for Mo films produced by physical vapor deposition. That data cube is available on the Materials Data Facility and describes 81 Mo films deposited with 27 unique process parameters and characterized by 8 techniques in triplicate or more for a total of 2072 raw datasets and 1889 quantities (e.g., film hardness, modulus) deduced from those datasets [38]. Building off that previous work, the current study explored 916 Ni-based electroplated films representing 160 unique deposition conditions, characterized across 6 modalities, resulting in a data cube consisting of 164,715 data files. These data have been made available in the Materials Data Facility, a public data repository [39].
Electroplated Ni films have long been used for wear/abrasion resistance, diffusion barriers in electrical connections, corrosion resistance, and improved visual appearance of coated surfaces [40]. Ni–Fe alloys find application in technologies requiring specialized magnetic functionalities, corrosion resistance, mechanical strength, and/or low coefficient of thermal expansion [41]. While extensive expertise has accumulated over nearly two centuries of electrochemical practice, the fundamentals connecting electrolyte composition and processing conditions to film performance are complex and not fully mastered. There is a myriad of process parameters in electrodeposition that can be optimized, including bath chemistry (metal salts, supporting electrolytes, pH buffers, acids or bases, brighteners, etc.) and plating conditions (temperature, current density, agitation, etc.) This expansive process space is well suited for exploration by an accelerated multimodal workflow. By repeating measurements for each individual sample to enhance statistical interpretation, the resulting data cube consisted of 164,715 unique data files under this accelerated workflow. This large dataset is amenable to future machine learning-based process exploration, and will be made available to the public in an open repository, employing FAIR data principles [42].
Methods
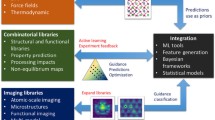
As illustrated in Fig. 1, Ni and Ni–Fe films were electrodeposited with a custom parallel plating system and evaluated with different characterization modalities. The data was input into a custom database (Experiment Tracker) to facilitate data transfer between our experimental and machine learning teams. Efforts to analyze the obtained data with PIMA [43], a physics-informed multimodal autoencoder, are on-going.
High-throughput workflow for characterizing electroplated films. After fabrication, the samples are mounted on either a universal or instrument-specific sample holder and characterized by four characterization modalities and two performance modalities. The data from each technique is loaded into a database from which scientists can access the data for machine learning. The output is process–structure–property knowledge that can be used to determine optimal conditions for a desired material property or function, such as electroplating Ni and Ni alloy films and optimizing for film hardness and friction
Electrodeposition of Ni and Ni–Fe Films
Ni and Ni–Fe films were produced under eight plating conditions (Table 1, condition 1 performed three times) and then characterized with six modalities. The Ni and Ni–Fe films were deposited using a custom-built potentiostat equipped with sixteen channels that run in parallel. Custom 3-electrode cells consisted of a Ni coiled wire counter electrode (Shopaid, ≥ 99% Ni), Ni wire pseudo-reference electrode (TE TEMCo, 99.6% Ni), and a Si working electrode coated with 100 nm Ti and 200 nm Au (e-beam evaporated). The working electrode area was defined by a 1 cm diameter Viton O-ring in a custom sample holder that isolated the sides of the Si/Ti/Au substrate from solution. Ni and Ni–Fe films were plated with eight different plating conditions (Table 1). All films were plated in 7 mL electrolyte baths consisting of 0.76 M nickel sulfate (NiSO4·6H2O, Fisher Scientific, Certified ACS) and 0.4 M boric acid (Fisher Chemical, Certified ACS). Four additional baths contained saccharin (Aldrich Chemical Co.) with concentrations of 0.06 mM, 0.5 mM, 2.7 mM, and 6 mM. Two series of films were produced with baths containing 13 mM FeSO4 (Acros Organics, puriss), one of which also contained 5 mM sodium lauryl sulfate (SLS, Aldrich, 99 + % ACS reagent grade) and 6 mM saccharin. All electrolytes were dissolved in deionized water. Experiments at constant current were performed with current densities between 1 and 200 mA cm−2 for durations intended to produce 1 μm thick films (assuming 100% current efficiency). Each deposited film was characterized with profilometry, X-ray diffraction (XRD), and X-ray fluorescence (XRF) at four sample locations. Point 1 is centered on the film and points 2–4 are offset 200 μm vertically and 200 μm horizontally from point 1. Optical microscopy was performed centered at point 1 as detailed below. Next, an 8 × 8 grid of indents centered on the electroplated area was created during nanoindentation. Friction testing was performed in three horizontal parallel tracks centered on the plated area. Figure 2 shows the data cube for measurements collected from 915 samples. Overall, electroplated samples were described with four characterization modalities and two performance metrics.
A data cube diagram illustrating the number of datasets generated from characterizing 915 electroplated samples. The samples were plated under 8 different plating conditions (Table 1, condition 1 repeated twice) and 20 current densities. Each unique current density and plating condition combination was replicated four or more times
Sample Storage and Labeling
After plating, films were stored in well plates (Corning, 12 wells, 3 mL each). A custom label for each sample was affixed to the well plate lid above the corresponding sample. Labels were designed using the Brother P-Touch Software and printed on a Brother QL-800 label printer. Labels included unique and formulaic sample numbers generated with a Sandia-developed database (Experiment Tracker), sample creation date, and a QR code encoding the sample number.
Optical Microscopy
After plating, optical micrographs were collected for each film using a Keyence VHX-7100. A total of 8 images were taken at sample point 1 for each film by varying the 20x vs. 100x lenses, coaxial vs ring lighting, and high dynamic range (HDR) vs standard imaging. The “Teaching Automatic Measurement” mode in the software was used to program sample locations and multiple lighting conditions for imaging with each objective lens. A custom 3D printed sample holder (Fig. 3a) enabled the saved “User Settings” to be used across multiple sample batches.
Laser Profilometry
Topographical height maps of the films were collected with a Keyence VK-X3100 Surface Profiler equipped with a 404 nm violet laser. The “Teaching Mode” in the instrument software was used to set the data collection parameters. We used python code to generate the data collection order, which allowed us to easily program the data collection locations at points 1–4. A micrograph and laser confocal height map of each sample were first collected at point 1 with the 20x objective lens then at points 1–4 with the apochromatic 150x objective lens. The collection order was incorporated into the “Teaching Mode” in the instrument software to enable the use of the motorized X–Y stage. A custom 3D printed sample holder (Fig. 3b) was used to load samples in a grid into the instrument. The “Teaching Mode” macro, software fiducial corrections, and custom sample holder enabled the same macro to be used across multiple sample batches without further modification. The Keyence MultiFileAnalyzer software Batch analysis function was used to flatten all the sample images with the “Reference plane settings” image processing tool. There is not a batch process to save the height maps and micrographs in the MultiFileAnalyzer software, so MacroRecorder software was used to create a macro of keyboard strokes and mouse clicks to automatically save csv files of the height maps and png files of the micrographs.
X-ray Diffraction (XRD)
XRD patterns of the plated films were collected with a Rigaku SmartLab II Powder XRD using Cu Kα radiation at 40 kV and 44 mA. A custom 3D printed holder (Fig. 3d) was affixed to the instrument X–Y motorized stage such that multiple samples could be analyzed in serial. The instrument software was used to program a macro that automatically performed a sample alignment for each row of samples and collected diffraction patterns for each sample. XRD patterns were collected at points 1–4 on each sample.
X-ray Fluorescence (XRF) Spectroscopy
XRF spectra of the films were collected with a Bruker M4 TORNADO with Rh Kα radiation at 40 kV and 300 μA. Each spectrum was collected with a 20 μm beam size for 30 s. The “mosaic” mode in the software was used to stitch together a map of all samples, and the “autopoint” mode was used to program the instrument to collect spectra at points 1–4 across all samples. A custom 3D printed sample holder (Fig. 3c) was used to load multiple samples into the instrument for automated characterization. The system energy calibration was confirmed using a Zr standard before data collection.
Nanoindentation
Nanoindentation was performed using a Bruker Hysitron TI980 nanoindenter with a custom magnetic stage plate with 16 sample locations. High-throughput nanoindentation was performed using high speed property mapping (XPM) [44] consisting of 64 indents per sample in a 8 × 8 grid with 9 μm spacing between indents (72 μm × 72 μm grid) to avoid overlap of indents. Load controlled indentation was performed with a Berkovich tip using a trapezoidal loading profile (0.1–0.5–0.1 s profile) with a max force of 1000 μN, resulting in indentation depths ranging from 30 nm–100 nm.
Friction Testing
The coefficient of friction was measured using a custom high-throughput ball-on-flat bidirectionally reciprocating tribometer capable of testing six samples in parallel (described elsewhere [45], with more details in the online Supplementary Material). Three tests were performed on each sample using a 1/8 inch diameter sapphire ball at a normal force of 50 mN for 500 cycles, a sliding speed of 4 mm/s, and track length of 2 mm. All tests were performed in a dry nitrogen environment (O2 < 200 ppm, H2O < 200 ppm).
Instrument-Specific Custom Sample Holders
Instrument-specific custom sample holders were designed in SolidWorks for optical microscopy, profilometry, XRF, and XRD (Fig. 3a–d). An Objet J826 printer (Stratasys, Edina, MN) was used to 3D print the holders in Stratasys photocurable materials (Vero™ White, Cyan, Black). The design of each sample holder was made to optimize the number of samples loaded into each instrument and includes ease of use features such as sample positioning guides and depressions to allow tweezers to easily reach under the samples. The number of samples supported by each instrument-specific holder was 45 for optical microscopy, 40 for laser profilometry, 31 for XRD, and 48 for XRF.
Universal Sample Holders with Instrument-Specific Adapters
A universal sample holder (Fig. 3e) supporting 20 samples for mounting into instrument-specific adapters was designed in SolidWorks. Adapters were fabricated to interface with individual instruments: XRD, XRF, profilometry, nanoindentation, and microscopy. An UltiMaker 3 Extended printer (UltiMaker, Utrecht, Netherlands) was used to 3D print the holders in polylactic acid (MatterHackers Inc., Lake Forest, CA).
Calculation of Time Savings
The time to load samples, acquire data, unload samples, and save data was recorded while an experienced operator analyzed 5 samples with and without improved workflow strategies implemented. Hands-on time for data acquisition includes the time to input parameters, align samples, and start automated data collection. Sample handling includes loading and unloading. Loading involves placing the samples either directly onto the instrument stage or onto a sample holder before placing the holder on the instrument stage. Unloading is the removal of samples from the instrument and placing them back into the well plate sample holder. The hands-on time to save data is the time taken to open data files and save them in a non-proprietary format. The time to save data was only recorded for profilometry because all other instruments and their software saved data in non-proprietary formats. While the instrument-specific sample holders were used to calculate time savings for each task, the time to perform these activities was normalized to 20 samples for each technique, equal to the number of samples mounted on a universal sample holder. Activities that did not incorporate accelerated workflow strategies were not timed.
Results
The goal of this study was to quantify efficiency gains with the streamlined/automated workflow over typical human-based artisan measurements. To assess workflow improvements, the different tasks associated with data acquisition, sample handling, and data saving for 20 samples were timed for each instrument. Figure 4a shows the hands-on time spent before workflow improvements were implemented and after the tasks were streamlined. The streamlined processes were 20x, 18x, 46x, and 13x faster for optical microscopy, profilometry, XRD, and XRF, respectively. Workflow acceleration strategies were not applied to nanoindentation or friction testing, so the values presented are estimates of the duration of manual and streamlined workflows. The time to perform each task before and after streamlining is tabulated in Table S1 and the time savings are in Table S2.
The largest hands-on time savings for all characterization methods originated from automating the data acquisition task (Fig. 4a) which was 110x, 30x, 226x, and 30x faster for optical microscopy, profilometry, XRD, and XRF, respectively. Greater improvements in hands-on time were observed for optical microscopy and XRD because XRF data acquisition was the fastest to perform manually (76 min) compared to the other techniques, and the profilometry data acquisition includes an additional fiduciary correction to ensure the macro is accurately mapped onto the current tray of samples. The accelerated characterization workflow resulted in the collection of a large multimodal dataset for 915 electroplated samples (Fig. 5).
a The amount of hands-on time spent per task before (manual) and after streamlined workflows strategies were implemented for 20 samples for data acquisition, saving data, and sample loading and unloading. The time to save data was only recorded for profilometry. The speedup estimates reported for nanoindentation and friction testing are anticipated with future employment of the universal holder. b The amount of hands-on time saved after implementing programming strategies to accelerate renaming files and uploading them into a database. The streamlined renaming of files takes 0 min of hands-on time because it is incorporated into the upload data task
Examples of data collected from each characterization modality. a Electrochemical voltage versus time curves that are zoomed in to show the first 20 s of the experiments. Some experiments lasted a duration of 1463.2 s. b Nanoindentation load curves for which 1/64 of the data available is visualized. c Coefficient of friction testing over 500 cycles for which 1/3 of the data available is shown. d A single XRD pattern to show the range from which the patterns in (e) originate. e XRD patterns of the relevant Fe and Au peaks for which 1/4 of the data available is shown. f A single XRF spectra to show the range from with the patterns in (g) originate. g XRF spectra showing the most prominent Ni and Fe peaks. The data shows 1/4 of the available data. h Optical microscopy (OM) of which 1/8 of the available images are shown. i Optical microscopy corresponding to j height maps collected with an optical profilometer. 1/5 of the available data is show in (i) and (j)
While each instrument had built-in automation functions, they were generally developed for within-sample exploration rather than between-sample exploration and were not optimized for high-throughput data acquisition of a grid of analysis locations on each sample. Specifically, the profilometer built-in macro writer was probably not intended to measure samples across the entire sample stage with the highest magnification lens on samples of slightly varying height. As a result, the instrument in its default configuration would not correctly autofocus on each sample resulting in missed datasets, so through software adjustments to maximize focus search range, the issue was mitigated. During sample registration, the lens was defocused and positioned approximately 110 μm closer to the sample compared to the in-focus position. This resulted in a larger accessible range during autofocus. Additionally, the sample acquisition order we utilized (multiple analysis locations per sample) would have been inefficient using the built-in grid tool, so we developed Python code to produce the most efficient sampling order. The XRD macro software cannot use embedded loops to execute tasks, so as a workaround, many repetitive tasks were listed in series resulting in a macro with 92 steps. Finally, the OM teaching program is robust in its ability to replicate tasks for which it has been “trained,” but editing that list of tasks is cumbersome. Training can take hours to generate the several hundred tasks required to image all samples on a holder under all lighting conditions and magnifications desired. To enable agile data collection under different conditions from samples varying in number, dimension, and arrangement, we developed a Python script to modify the tasks easily without manual retraining. The script writes task directions and then builds the library of reference images into the training folder which allows any quantity of images to be taken under any conditions.
Custom 3D printed sample holders aided in reducing the hands-on time for data acquisition and sample loading and unloading. Holders that consistently allow samples to be placed in the same position on a sample stage enable the use of instrument macros for data acquisition. Without a sample holder, automation can still be used but the operator would have to carefully align samples in a grid and measure sample locations to ensure the macro would collect data from the correct location on the sample. The use of a custom instrument-specific holder accelerated sample loading and unloading by 1.0x–3.5x. An operator can move samples a short distance from the sample storage container to the sample holder nearby faster than they can transfer a sample into the instrument. Additionally, guides on the sample holders allow for fast alignment of samples compared to manual manipulation of samples on the instrument stage with tweezers. The greatest improvement in loading time was recorded for XRD, which includes the time to measure the sample locations on the instrument stage because the instrument relies on moving the stage to set coordinates without the aid of a camera. For all other techniques, samples were manually loaded in an approximate grid and then navigated to with the aid of a camera. To further accelerate sample loading and unloading, custom sample holders were designed with depressions for tweezer insertion under the samples.
Custom sample holders accelerate workflow by ensuring consistent sample alignment and uniform sample spacing. Two different holder strategies were explored: single sample holders per instrument (Fig. 3a–d) or universal sample holders (Fig. 3e) paired with instrument-specific adapters (Fig. 3f). The use of a single instrument-specific custom sample holder is a resource efficient method to speed up all hands-on tasks related to sample characterization. Samples are transported to an instrument and loaded onto the instrument-specific holder, analyzed, unloaded, and then carried to the next instrument. A more material expensive strategy to further accelerate sample handling is to use a universal sample holder that fits into instrument-specific adapters. Samples are loaded onto the universal holder once, and that holder is carried to each instrument and inserted into an instrument-specific adapter. A universal sample holder essentially eliminates all loading and unloading times. As a trade-off, the universal holder must function in every instrument, so the number of samples loaded into the instrument may be diminished by accommodating the smallest instrument stage in a multimodal workflow. For ease of programming across all platforms, we chose to design a universal holder with a rectangular array of samples. The XRD utilized has the smallest stage, and so limited the number of samples (20) for a universal holder. Additional considerations, such as the relatively large XRF stage size, resulted in designing a stage adapter that could fit 4 units of the universal holder so appreciable numbers of samples could be maximized.
Making sure our data outputs were in accessible and interoperable formats was key to ensuring our data was FAIR [42]. As such, we developed a strategy to save the proprietary profilometry data in non-proprietary, releasable file formats. The profilometry data proprietary format requires the user to employ the instrument manufacturer’s data software to open the file and subsequently export the image data as png files and the height maps as csv files. Since there is not a batch functionality to save the data, a mouse-and-keyboard-stroke recorder (MacroRecorder) was used to program a sequence to save up to ~ 200 datasets once uploaded into the image processing (MultiFileAnalyzer) software. Figure 4b shows the streamlined process where the MacroRecorder saved 32 min for 20 samples and was 64x faster than saving the data by hand. All other instruments were capable of saving the raw data in non-proprietary file formats and so time improvements for data saving were not relevant.
Most instruments were built with the intention that a human operator would manually name each file. Even with automation, the naming convention typically was only a numerical increment on a base file name. Such systems are adequate for collecting numerous nominally identical observations on a single sample, but are inadequate for multiple samples (with repeated observations within each sample). Appropriately naming the data files with unique and succinct identifiers was not easily achievable on any instrument while also utilizing the built-in high-throughput functionalities. All of the instruments would increment file names, and so Python code was developed to rename all of the files based on the folder name. The folder name listed the range of numerical sample identifiers then stepped through the files and renamed them in order. On average, it takes an operator 2.2 min to change the names of 20 files. For our experiments, 20 characterization files originated from characterizing a single sample with OM, profilometry, XRD, and XRF. Therefore, renaming all files for a tray of 20 samples would take 44 min. The code developed to rename files was incorporated into the database upload code discussed below, so the amount of hands-on time saved per sample was 44 min (Fig. 3b).
Producing and sharing large numbers of samples and their characterization data with a delocalized group of collaborators led our team to develop a database (Experiment Tracker) to organize, tag, and track all samples and related data. Manually uploading data and relevant metadata about process and characterization conditions takes approximately 35 min per sample when four analysis locations for four characterization methods were performed (Fig. 3b). An implemented alternative was to employ Experiment Tracker’s application programming interface (API) to automate the process. The implemented code automatically and periodically scans a shared folder for new data additions then renames files, extracts relevant metadata, uploads all data, and updates the database. The benefits of using this API approach include: hands-on time to upload data is limited to the time to copy and paste data into the correct directory, data upload and metadata updates are performed in the background, the approach is scalable to support different characterization modalities, mistakes in typing metadata keywords and associated values are avoided, steps in the process are not forgotten (e.g., user forgets to upload data after updating metadata), and data upload can be automatically verified. While development time for coding was not tracked, the time savings are apparent. For 1000 samples, a trained scientist would spend 24 days uploading data and related metadata, an unacceptable cost for any high-throughput project hoping to produce hundreds to thousands of samples.
Discussion
We have shown strategies to accelerate metallic film characterization workflows, including strategically naming and tracking samples, utilizing embedded instrument automation features, taking advantage of instrument mapping functionality via custom sample holders, and utilizing a click-and-keyboard-stroke macro builder to overcome instrument automation limitations. These relatively easy-to-implement, low-cost strategies support the collection of large materials science datasets (Fig. 5) to aid in machine learning, increase the robustness of artisanal experimentation through improved statistics, and allow for wider experimentation within a materials design space. They differ from other automation efforts that utilize extensively customized robotic systems [26, 27, 37].
The approach taken in the current study is differentiated from other accelerated characterization research due to its applicability to almost any instrument with pre-existing automation features. In the current study, significant time savings were accomplished by employing (1) pre-existing automation features, (2) strategies to overcome instrument limitations, and (3) custom sample holders. Unlike fully automated workflows that use robotics to transfer samples between various synthesis/characterization stations [26, 27, 37], the substantial speedups demonstrated herein are broadly transferrable to other domains/modalities, often with little-to-no capital investment and minimal customization. Consequentially, the methodology presented can be implemented within almost any size laboratory and for a wide array of applications [38]. In contrast, many prior works have provided automated or accelerated workflows that are not easily transferrable across modalities or sample types [12]. Furthermore, automation using robotics often requires that all instruments are in physical proximity whereas the current approach can be adopted when individual instruments are located in different rooms, buildings, or facilities.
An additional consideration for acceleration of multiple instruments in a workflow is throughput-matching or throughput management. In the current approach, we selected characterization modalities that were amenable to ~ 50–100 measurements per day. By handling the samples in multi-sample holders, an entire rack of specimens could be analyzed within a day. In this way, each modality took no more than a day to complete, and the entire workflow could be completed in 1–2 weeks. An additional workflow consideration was the time required to shuttle batches of samples between instruments and coordination between multiple operators. While not utilized in the present study, there are software solutions for workflow tracking and coordination, commonly used in manufacturing environments.
An accelerated materials characterization workflow complements techniques that are already considered to be high throughput by enabling more diverse multimodal data to be collected for each sample. For example, nanoindentation has been considered a high-throughput technique compared to other materials property techniques for over two decades [44, 46,47,48]. Our workflow enables the high-throughput collection of additional information on sample composition, structure, and morphology through nondestructive modalities to enrich the high-throughput mechanical knowledge derived from nanoindentation. Overall, a high-throughput workflow facilitates the collection of more holistic datasets describing a collection of samples.
The presented flexible and adaptable approach to workflow automation enables the reduction of human error and bias during data collection. Human error occurred in our process development while an experienced researcher manually named files and mis-navigated to the specific analysis locations on the samples. The streamlined workflows use automation via mapping to navigate to analysis locations and programming with Python to automate file renaming to avoid common operator mistakes.
The cost savings achieved by adopting a high-throughput characterization workflow for many samples are significant. As a ballpark estimate, assume a full-time scientist with overhead costs a project $400,000 per year or $192 per hour. The times savings presented herein for performing optical microscopy, profilometry, XRD, and XRF on 915 samples yields a cost savings of over $66,000 for the materials characterization and $95,000 for the data upload. This translates into two person months of savings. Those person months of an expert scientist can be translated into deeper data analysis and interpretation.
Summary and Conclusions
High-throughput characterization workflows can support wider sampling of fabrication variable spaces, statistical treatment of data enabled by multiple replicates, multi-variate regression, machine learning, support vector machines, and other methods. Task automation within high-throughput workflows is a key investment needed to accelerate multimodal characterization. It can minimize human errors, lower costs, and allow scientists to allocate their time to critical thinking as opposed to repetitive manual tasks. Herein, we describe an implemented workflow for accelerating the characterization of thin films. The acceleration strategies do not require manufacturing-scale resources and are thus amenable to typical materials research facilities in academic, government, or commercial laboratories. To avoid building custom instrumentation, the workflow leverages existing instrumentation and augments their weaknesses with time-saving strategies that can be implemented in whole or part by others, dependent on project needs and budget. The streamlined processes required 20x, 18x, 46x, and 13x less hands-on time than manually characterizing samples for optical microscopy, profilometry, XRD, and XRF, respectively. Furthermore, we project that the streamlined workflow strategies will accelerate nanoindentation and friction testing by ~ 10x in future studies.
References
Mansouri Tehrani A, Oliynyk AO, Parry M, Rizvi Z, Couper S, Lin F, Miyagi L, Sparks TD, Brgoch J (2018) Machine learning directed search for ultraincompressible, superhard materials. J Am Chem Soc 140(31):9844–9853
Oliynyk AO, Adutwum LA, Harynuk JJ, Mar A (2016) Classifying crystal structures of binary compounds AB through cluster resolution feature selection and support vector machine analysis. Chem Mater 28(18):6672–6681
Wahl CB, Aykol M, Swisher JH, Montoya JH, Suram SK, Mirkin CA (2021) Machine learning–accelerated design and synthesis of polyelemental heterostructures. Sci Adv 7(52):eabj5505
Belsky A, Hellenbrandt M, Karen VL, Luksch P (2002) New developments in the inorganic crystal structure database (ICSD): accessibility in support of materials research and design. Acta Crystallogr Sect B Struct Sci 58(3):364–369
Zakutayev A, Wunder N, Schwarting M, Perkins JD, White R, Munch K, Tumas W, Phillips C (2018) An open experimental database for exploring inorganic materials. Sci Data 5(1):1–12
Curtarolo S, Setyawan W, Hart GL, Jahnatek M, Chepulskii RV, Taylor RH, Wang S, Xue J, Yang K, Levy O (2012) AFLOW: an automatic framework for high-throughput materials discovery. Comput Mater Sci 58:218–226
Jain A, Ong SP, Hautier G, Chen W, Richards WD, Dacek S, Cholia S, Gunter D, Skinner D, Ceder G (2013) Commentary: the materials project: a materials genome approach to accelerating materials innovation. APL Mater 1(1):011002
Kirklin S, Saal JE, Meredig B, Thompson A, Doak JW, Aykol M, Rühl S, Wolverton C (2015) The open quantum materials database (OQMD): assessing the accuracy of DFT formation energies. NPJ Comput Mater 1(1):1–15
Allen FH (2002) The cambridge structural database: a quarter of a million crystal structures and rising. Acta Crystallogr Sect B Struct Sci 58(3):380–388
Watson C (2022) Many researchers say they’ll share data—but don’t. Nature 606(7916):853–853
McFarland EW, Weinberg WH (1999) Combinatorial approaches to materials discovery. Trends Biotechnol 17(3):107–115
Maier WF, Stoewe K, Sieg S (2007) Combinatorial and high-throughput materials science. Angew Chem Int Ed 46(32):6016–6067
Reizman BJ, Wang Y-M, Buchwald SL, Jensen KF (2016) Suzuki–Miyaura cross-coupling optimization enabled by automated feedback. React Chem Eng 1(6):658–666
McMullen JP, Jensen KF (2010) An automated microfluidic system for online optimization in chemical synthesis. Org Process Res Dev 14(5):1169–1176
Grasser JA, Muggli DS (2009) A high-throughput reaction system to measure the gas-phase photocatalytic oxidation activity of TiO2 nanotubes. Rev Sci Instrum 80(7):075106
Wollmann P, Leistner M, Stoeck U, Grünker R, Gedrich K, Klein N, Throl O, Grählert W, Senkovska I, Dreisbach F (2011) High-throughput screening: speeding up porous materials discovery. Chem Commun 47(18):5151–5153
Gregoire JM, Xiang C, Liu X, Marcin M, Jin J (2013) Scanning droplet cell for high throughput electrochemical and photoelectrochemical measurements. Rev Sci Instrum 84(2):024102
Sliozberg K, Schäfer D, Erichsen T, Meyer R, Khare C, Ludwig A, Schuhmann W (2015) High-throughput screening of thin-film semiconductor material libraries I: system development and case study for Ti–W–O. Chemsuschem 8(7):1270–1278
Medina-Franco JL, Sanchez-Cruz N, Lopez-Lopez E, Diaz-Eufracio BI (2022) Progress on open chemoinformatic tools for expanding and exploring the chemical space. J Comput Aided Mol Des 36(5):341–354
Menon A, Childs CM, Poczós B, Washburn NR, Kurtis KE (2019) Molecular engineering of superplasticizers for metakaolin-portland cement blends with hierarchical machine learning. Adv Theory Simul 2(4):1800164
Bucior BJ, Bobbitt NS, Islamoglu T, Goswami S, Gopalan A, Yildirim T, Farha OK, Bagheri N, Snurr RQ (2019) Energy-based descriptors to rapidly predict hydrogen storage in metal–organic frameworks. Mol Syst Des Eng 4(1):162–174
Ling J, Hutchinson M, Antono E, Paradiso S, Meredig B (2017) High-dimensional materials and process optimization using data-driven experimental design with well-calibrated uncertainty estimates. Integr Mater Manuf Innov 6:207–217
Raccuglia P, Elbert KC, Adler PD, Falk C, Wenny MB, Mollo A, Zeller M, Friedler SA, Schrier J, Norquist AJ (2016) Machine-learning-assisted materials discovery using failed experiments. Nature 533(7601):73–76
Ren F, Ward L, Williams T, Laws KJ, Wolverton C, Hattrick-Simpers J, Mehta A (2018) Accelerated discovery of metallic glasses through iteration of machine learning and high-throughput experiments. Sci Adv 4(4):eaaq1566
Saal JE, Oliynyk AO, Meredig B (2020) Machine learning in materials discovery: confirmed predictions and their underlying approaches. Annu Rev Mater Res 50:49–69
MacLeod BP, Parlane FG, Morrissey TD, Häse F, Roch LM, Dettelbach KE, Moreira R, Yunker LP, Rooney MB, Deeth JR (2020) Self-driving laboratory for accelerated discovery of thin-film materials. Sci. Adv 6(20):eaaz8867
MacLeod BP, Parlane FG, Brown AK, Hein JE, Berlinguette CP (2022) Flexible automation accelerates materials discovery. Nat Mater 21(7):722–726
Dieckhöfer S, Schuhmann W, Ventosa E (2021) Accelerated electrochemical investigation of Li plating efficiency as key parameter for Li metal batteries utilizing a scanning droplet cell. ChemElectroChem 8(16):3143–3149
Epps RW, Bowen MS, Volk AA, Abdel-Latif K, Han S, Reyes KG, Amassian A, Abolhasani M (2020) Artificial chemist: an autonomous quantum dot synthesis bot. Adv Mater 32(30):2001626
Guevarra D, Haber JA, Wang Y, Zhou L, Kan K, Richter MH, Gregoire JM (2022) High throughput discovery of complex metal oxide electrocatalysts for the oxygen reduction reaction. Electrocatalysis 13:1–10
Hitt JL, Yoon D, Shallenberger JR, Muller DA, Mallouk TE (2022) High-throughput fluorescent screening and machine learning for feature selection of electrocatalysts for the alkaline hydrogen oxidation reaction. ACS Sustain Chem Eng 10(49):16299–16312
Joress H, DeCost B, Hassan N, Braun TM, Gorham JM, Hattrick-Simpers J (2022) Development of an automated millifluidic platform and data-analysis pipeline for rapid electrochemical corrosion measurements: a pH study on Zn–Ni. Electrochim Acta 428:140866
Long C, Bunker D, Li X, Karen V, Takeuchi I (2009) Rapid identification of structural phases in combinatorial thin-film libraries using x-ray diffraction and non-negative matrix factorization. Rev Sci Instrum 80(10):103902
Mitrovic S, Cornell EW, Marcin MR, Jones RJ, Newhouse PF, Suram SK, Jin J, Gregoire JM (2015) High-throughput on-the-fly scanning ultraviolet-visible dual-sphere spectrometer. Rev Sci Instrum 86(1):013904
Takeuchi I, Long C, Famodu O, Murakami M, Hattrick-Simpers J, Rubloff G, Stukowski M, Rajan K (2005) Data management and visualization of x-ray diffraction spectra from thin film ternary composition spreads. Rev Sci Instrum 76(6):062223
Zhou L, Guevarra D, Gregoire JM (2022) High throughput discovery of enhanced visible photoactivity in Fe–Cr vanadate solar fuels photoanodes. J Phys Energy 4(4):044001
Burger B, Maffettone PM, Gusev VV, Aitchison CM, Bai Y, Wang X, Li X, Alston BM, Li B, Clowes R (2020) A mobile robotic chemist. Nature 583(7815):237–241
Kalaswad M, Custer J, Addamane S, Khan R, Jauregui L, Babuska T, Henriksen A, DelRio F, Dingreville R, Boyce B (2023) Sputter-deposited Mo Thin films: multimodal characterization of structure, surface morphology, density, residual stress, electrical resistivity, and mechanical response. Integr Mater Manuf Innov 12:1–12
Bassett KL, Watkins T, Coleman J, Bianco N, Bailey LS, Pillars J, Williams SG, Babuska TF, Curry J, DelRio FW, Henriksen AA, Garland A, Hall J, Krick BA, Boyce BL (2023) Accelerated multimodal characterization of Ni and Ni-Fe electrodeposited films. Mater Data Facil. https://doi.org/10.18126/V84F-WE14
Mahidashti Z, Aliofkhazraei M, Lotfi N (2018) Review of nickel-based electrodeposited tribo-coatings. Trans Indian Inst Met 71:257–295
Lin Q, Hoglund E, Zangari G (2020) Electrodeposition of Fe–Ni alloy on Au(111) substrate: metastable BCC growth via hydrogen evolution and interactions. Electrochimica Acta 338:135876
Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten J-W, da Silva Santos LB, Bourne PE (2016) The FAIR guiding principles for scientific data management and stewardship. Sci Data 3(1):1–9
Trask N, Martinez C, Lee K, Boyce B (2022) Unsupervised physics-informed disentanglement of multimodal data for high-throughput scientific discovery. arXiv preprint arXiv:2202.03242
Hintsala ED, Hangen U, Stauffer DD (2018) High-throughput nanoindentation for statistical and spatial property determination. JOM 70(4):494–503
Nation BL, Dugger MT, Curry J (2022) High throughput tribometer. US 2022/0107254 A1
Warren OL, Wyrobek TJ (2004) Nanomechanical property screening of combinatorial thin-film libraries by nanoindentation. Meas Sci Technol 16(1):100
Miracle DB, Li M, Zhang Z, Mishra R, Flores KM (2021) Emerging capabilities for the high-throughput characterization of structural materials. Annu Rev Mater Res 51:131–164
Liu Y, Zhang X (2018) Beyond indentation hardness and modulus: recent advances in nanoindentation techniques: part II. JOM 70(4):485–486
Acknowledgements
The authors were supported by a Laboratory Directed Research and Development (LDRD) program. This work was performed, in part, at the Center for Integrated Nanotechnologies, an Office of Science User Facility operated for the US Department of Energy (DOE) Office of Science. Sandia National Laboratories is a multimission laboratory managed and operated by National Technology & Engineering Solutions of Sandia, LLC, a wholly owned subsidiary of Honeywell International, Inc., for the US DOE’s National Nuclear Security Administration under contract DE-NA-0003525. The views expressed in the article do not necessarily represent the views of the US DOE or the US Government.
Funding
Sandia National Laboratories.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bassett, K.L., Watkins, T., Coleman, J. et al. A Workflow for Accelerating Multimodal Data Collection for Electrodeposited Films. Integr Mater Manuf Innov 12, 430–440 (2023). https://doi.org/10.1007/s40192-023-00315-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40192-023-00315-5