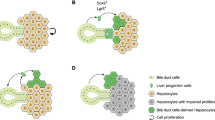
Abstract
The regenerative capability of the liver is essential for recovery from all hepatic injuries. Although long studied, the signals that regulate such regeneration require further elucidation if knowledge about regenerative mechanisms is to be translated into improved clinical therapy. Alterations in metabolism have been the focus of recent experimental investigations as a possible source of essential signals that control liver regeneration. Although the specific mechanisms linking metabolism and regeneration remain unknown, specific growth factors, secondary messengers, and transcription factors have been suggested by published analyses. Epigenetic mechanisms are also emerging as potential intermediaries between hepatic insufficiency-induced changes in metabolism and regenerative hepatocellular proliferation. This article reviews the recent literature relevant to these considerations, with particular emphasis on contemporary data that link metabolic and epigenetic signals to the regulation of liver regulation. The relevance of metabolic–epigenetic regulation of experimental hepatic regeneration with respect to human liver diseases is also briefly considered.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Davenport RJ (2005) What controls organ regeneration? Science 309(5731):84
• Rudnick DA (2014) Liver regeneration: the developmental biologists approach. In: Orlando G, Lerut JP, Soker S, Stratta RJ (eds) Regenerative medicine applications in organ transplantation, 1st edn. Elsevier/Academic Press, Waltham, pp 353–374. This recent chapter provides an overview of the history of experimental analyses of liver regeneration and the model systems employed for and broad principles and specific molecular regulators elucidated by such regulation
• Agathocleous M, Harris WA (2013) Metabolism in physiological cell proliferation and differentiation. Trends Cell Biol 23(10):484–492. This review considers the data in support of and mechanisms by which metabolism is linked to cell proliferation
• Ward PS, Thompson CB (2012) Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 21(3):297–308. This review considers the data in support of metabolic reprogramming as promoting oncogenic cell proliferation
Vander Heiden MG, Cantley LC, Thompson CB (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324(5930):1029–1033
Rudnick DA, Davidson NO (2012) Functional relationships between lipid metabolism and liver regeneration. Int J Hepatol 26:2012
•• Huang J, Rudnick DA (2014) Elucidating the metabolic regulation of liver regeneration. Am J Pathol 184(2):309–321. This recent review provides an overview of recent and previously published data in support of a metabolic model of liver regeneration and considers the clinical implications and areas for further study suggested by such findings
Weymann A, Hartman E, Gazit V, Wang C, Glauber M, Turmelle Y et al (2009) p21 is required for dextrose-mediated inhibition of mouse liver regeneration. Hepatology 19(50):207–215
• Mehendale HM (2005) Tissue repair: an important determinant of final outcome of toxicant-induced injury. Toxicol Pathol 33(1):41–51. This paper reviews the mechanisms that regulate liver repair after toxin-induced injury, including nutritional modifiers of toxin-induced liver regeneration
• Fanti M, Singh S, Ledda-Columbano GM, Columbano A, Monga SP (2014) Tri-iodothyronine induces hepatocyte proliferation by protein kinase A-dependent beta-catenin activation in rodents. Hepatology 59(6):2309–2320. This paper reports evidence supporting the role of PKA-dependent activation of β-catenin in T3-stimulated hepatocellular proliferation
Rudnick DA, Perlmutter DH, Muglia LJ (2001) Prostaglandins are required for CREB activation and cellular proliferation during liver regeneration. Proc Natl Acad Sci USA 98(15):8885–8890
Rudnick DA, Muglia LJ (2004) Eicosanoids and liver regeneration. In: Curtis-Prior Peter (ed) The Eicosanoids. Wiley, West Sussex, pp 415–422
Goessling W, North TE, Loewer S, Lord AM, Lee S, Stoick-Cooper CL et al (2009) Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell 136(6):1136–1147
De CD, Fimia GM, Sassone-Corsi P (1999) Signaling routes to CREM and CREB: plasticity in transcriptional activation. Trends Biochem Sci 24(7):281–285
Thompson MD, Monga SP (2007) WNT/beta-catenin signaling in liver health and disease. Hepatology 45(5):1298–1305
Chen H, Yang S, Yang Z, Ma L, Jiang D, Mao J et al (2007) Inhibition of GSK-3beta decreases NF-kappaB-dependent gene expression and impairs the rat liver regeneration. J Cell Biochem 102(5):1281–1289
Sekiya S, Suzuki A (2011) Glycogen synthase kinase 3 beta-dependent Snail degradation directs hepatocyte proliferation in normal liver regeneration. Proc Natl Acad Sci USA 108(27):11175–11180
Jin J, Wang GL, Shi X, Darlington GJ, Timchenko NA (2009) The age-associated decline of glycogen synthase kinase 3beta plays a critical role in the inhibition of liver regeneration. Mol Cell Biol 29(14):3867–3880
Jensen J, Brennesvik EO, Lai YC, Shepherd PR (2007) GSK-3beta regulation in skeletal muscles by adrenaline and insulin: evidence that PKA and PKB regulate different pools of GSK-3. Cell Signal 19(1):204–210
Huang W, Ma K, Zhang J, Qatanani M, Cuvillier J, Liu J et al (2006) Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science 312(5771):233–236
Meng Z, Wang Y, Wang L, Jin W, Liu N, Pan H et al (2010) FXR regulates liver repair after CCl4-induced toxic injury. Mol Endocrinol 24(5):886–897
Zhang L, Wang YD, Chen WD, Wang X, Lou G, Liu N et al (2012) Promotion of liver regeneration/repair by farnesoid X receptor in both liver and intestine in mice. Hepatology 56(6):2336–2343
• Borude P, Edwards G, Walesky C, Li F, Ma X, Kong B et al (2012) Hepatocyte-specific deletion of farnesoid X receptor delays but does not inhibit liver regeneration after partial hepatectomy in mice. Hepatology 56(6):2344–2352. These papers implicate hepatic and intestinal FXR expression as important regulators of liver regeneration
Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG et al (2005) Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab 2(4):217–225
Uriarte I, Fernandez-Barrena MG, Monte MJ, Latasa MU, Chang HC, Carotti S et al (2013) Identification of fibroblast growth factor 15 as a novel mediator of liver regeneration and its application in the prevention of post-resection liver failure in mice. Gut 62:899–910
Kong B, Huang J, Zhu Y, Li G, Williams J, Shen S et al (2014) Fibroblast growth factor 15 deficiency impairs liver regeneration in mice. Am J Physiol Gastrointest Liver Physiol 306(10):G893–G902
Naugler WE (2014) Bile acid flux is necessary for normal liver regeneration. PLoS ONE 9(5):e97426
Meng Z, Liu N, Fu X, Wang X, Wang YD, Chen WD et al (2011) Insufficient bile acid signaling impairs liver repair in CYP27(−/−) mice. J Hepatol 55:885–895
Pean N, Doignon I, Garcin I, Besnard A, Julien B, Liu B et al (2013) The receptor TGR5 protects the liver from bile acid overload during liver regeneration in mice. Hepatology 58(4):1451–1460
Fernandez-Barrena MG, Monte MJ, Latasa MU, Uriarte I, Vicente E, Chang HC et al (2012) Lack of Abcc3 expression impairs bile-acid induced liver growth and delays hepatic regeneration after partial hepatectomy in mice. J Hepatol 56(2):367–373
Manley S, Ni HM, Kong B, Apte U, Guo G, Ding WX (2014) Suppression of autophagic flux by bile acids in hepatocytes. Toxicol Sci 137(2):478–490
• Toshima T, Shirabe K, Fukuhara T, Ikegami T, Yoshizumi T, Soejima Y et al (2014) Suppression of autophagy during liver regeneration impairs energy charge and hepatocyte senescence in mice. Hepatology 60(1):290–300. This study reports data indicating that autophagy plays a critical role in liver regeneration
Amaya MJ, Oliveira AG, Guimaraes ES, Casteluber MC, Carvalho SM, Andrade LM et al (2014) The insulin receptor translocates to the nucleus to regulate cell proliferation in liver. Hepatology 59(1):274–283
Newberry EP, Kennedy SM, Xie Y, Luo J, Stanley SE, Semenkovich CF et al (2008) Altered hepatic triglyceride content after partial hepatectomy without impaired liver regeneration in multiple murine genetic models. Hepatology 48(4):1097–1105
Kohjima M, Tsai TH, Tackett BC, Thevananther S, Li L, Chang BH et al (2013) Delayed liver regeneration after partial hepatectomy in adipose differentiation related protein-null mice. J Hepatol 59(6):1246–1254
Obayashi Y, Campbell JS, Fausto N, Yeung RS (2013) Impaired lipid accumulation in the liver of Tsc2-heterozygous mice during liver regeneration. Biochem Biophys Res Commun 437(1):146–150
Ben YA, Lalazar G, Zolotaryova L, Steinhardt Y, Lichtentein Y, Ilan Y et al (2013) Impaired liver regeneration by beta-glucosylceramide is associated with decreased fat accumulation. J Dig Dis 14(8):425–432
Pauta M, Rotllan N, Vales F, Fernandez-Hernando A, Allen RM, Ford DA et al (2013) Impaired liver regeneration in Ldlr−/− mice is associated with an altered hepatic profile of cytokines, growth factors, and lipids. J Hepatol 59(4):731–737
Chakravarthy MV, Lodhi IJ, Yin L, Malapaka RR, Xu HE, Turk J et al (2009) Identification of a physiologically relevant endogenous ligand for PPAR alpha in liver. Cell 138(3):476–488
Gazit V, Huang J, Weymann A, Rudnick DA (2012) Analysis of the role of hepatic PPAR gamma expression during mouse liver regeneration. Hepatology 56(4):1489–1498
Turmelle YP, Shikapwashya O, Tu S, Hruz PW, Yan Q, Rudnick DA (2006) Rosiglitazone inhibits mouse liver regeneration. FASEB J 20:2609–2611
Lo Sasso G, Celli N, Caboni M, Murzilli S, Salvatore L, Morgano A et al (2009) Down-regulation of the LXR transcriptome provides the requisite cholesterol levels to proliferating hepatocytes. Hepatology 51:1334–1344
Mitro N, Mak PA, Vargas L, Godio C, Hampton E, Molteni V et al (2007) The nuclear receptor LXR is a glucose sensor. Nature 445(7124):219–223
Crumm S, Cofan M, Juskeviciute E, Hoek JB (2008) Adenine nucleotide changes in the remnant liver: an early signal for regeneration after partial hepatectomy. Hepatology 48(3):898–908
Vazquez-Chantada M, Ariz U, Varela-Rey M, Embade N, Martinez-Lopez N, Fernandez-Ramos D et al (2009) Evidence for LKB1/AMP-activated protein kinase/endothelial nitric oxide synthase cascade regulated by hepatocyte growth factor, S-adenosylmethionine, and nitric oxide in hepatocyte proliferation. Hepatology 49(2):608–617
• Merlen G, Gentric G, Celton-Morizur S, Foretz M, Guidotti JE, Fauveau V et al (2014) AMPKalpha1 controls hepatocyte proliferation independently of energy balance by regulating Cyclin A2 expression. J Hepatol 60(1):152–159. These papers report data supporting the role of AMPK in the regulation of liver regeneration
Hay N, Sonenberg N (2004) Upstream and downstream of mTOR. Genes Dev 18(16):1926–1945
Nelsen CJ, Rickheim DG, Tucker MM, Hansen LK, Albrecht JH (2003) Evidence that cyclin D1 mediates both growth and proliferation downstream of TOR in hepatocytes. J Biol Chem 278(6):3656–3663
Goggin MM, Nelsen CJ, Kimball SR, Jefferson LS, Morley SJ, Albrecht JH (2004) Rapamycin-sensitive induction of eukaryotic initiation factor 4F in regenerating mouse liver. Hepatology 40(3):537–544
Espeillac C, Mitchell C, Celton-Morizur S, Chauvin C, Koka V, Gillet C et al (2011) S6 kinase 1 is required for rapamycin-sensitive liver proliferation after mouse hepatectomy. J Clin Invest 121(7):2821–2832
Jakobsen JS, Waage J, Rapin N, Bisgaard HC, Larsen FS, Porse BT (2013) Temporal mapping of CEBPA and CEBPB binding during liver regeneration reveals dynamic occupancy and specific regulatory codes for homeostatic and cell cycle gene batteries. Genome Res 23(4):592–603
Fuchs O (2007) Growth-inhibiting activity of transcription factor C/EBPalpha, its role in haematopoiesis and its tumour suppressor or oncogenic properties in leukaemias. Folia Biol (Praha) 53(3):97–108
• Jin J, Hong IH, Lewis K, Iakova P, Breaux M, Jiang Y et al (2014) Cooperation of C/EBP family proteins and chromatin remodeling proteins is essential for termination of liver regeneration. Hepatology 2014 61(1):315–325. This paper provides evidence for the essential role of Cebpa in the termination of liver regeneration, which suggests down-regulation of Cebpa contributes to initiation of regeneration
Kurinna S, Stratton SA, Tsai WW, Akdemir KC, Gu W, Singh P et al (2010) Direct activation of forkhead box O3 by tumor suppressors p53 and p73 is disrupted during liver regeneration in mice. Hepatology 52(3):1023–1032
• Kurinna S, Stratton SA, Coban Z, Schumacher JM, Grompe M, Duncan AW et al (2013) p53 regulates a mitotic transcription program and determines ploidy in normal mouse liver. Hepatology 2013 57(5):2004–2013. This paper characterizes PH-induced liver regeneration in p53 null mice
Huang J, Glauber M, Qiu Z, Gazit V, Dietzen DJ, Rudnick DA (2012) The influence of skeletal muscle on the regulation of liver:body mass and liver regeneration. Am J Pathol 180(2):575–582
Paine-Saunders S, Viviano BL, Zupicich J, Skarnes WC, Saunders S (2000) glypican-3 controls cellular responses to Bmp4 in limb patterning and skeletal development. Dev Biol 225(1):179–187
Liu B, Paranjpe S, Bowen WC, Bell AW, Luo JH, Yu YP et al (2009) Investigation of the role of glypican 3 in liver regeneration and hepatocyte proliferation. Am J Pathol 175(2):717–724
Liu B, Bell AW, Paranjpe S, Bowen WC, Khillan JS, Luo JH et al (2010) Suppression of liver regeneration and hepatocyte proliferation in hepatocyte-targeted glypican 3 transgenic mice. Hepatology 52(3):1060–1067
Yang H, Xiong Y, Guan K (2013) Metabolic alteration in tumorigenesis. Sci China Life Sci 56(12):1067–1075
• Huang J, Barr E, Rudnick DA (2013) Characterization of the regulation and function of zinc-dependent histone deacetylases during rodent liver regeneration. Hepatology 57(5):1742–1751. This paper characterizes the regulation and functional importance of Zn-HDACs during liver regeneration
Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB (2009) ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324(5930):1076–1080
•• Rathmell JC, Newgard CB (2009) Biochemistry. A glucose-to-gene link. Science 324(5930):1021–1022. These papers link glucose metabolism to protein acetylation
Xia J, Zhou Y, Ji H, Wang Y, Wu Q, Bao J et al (2013) Loss of histone deacetylases 1 and 2 in hepatocytes impairs murine liver regeneration through Ki67 depletion. Hepatology 58(6):2089–2098
Ke Q, Yang RN, Ye F, Wang YJ, Wu Q, Li L et al (2012) Impairment of liver regeneration by the histone deacetylase inhibitor valproic acid in mice. J Zhejiang Univ Sci B 13(9):695–706
Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG et al (2011) Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell 145(4):607–621
Ha CH, Kim JY, Zhao J, Wang W, Jhun BS, Wong C et al (2010) PKA phosphorylates histone deacetylase 5 and prevents its nuclear export, leading to the inhibition of gene transcription and cardiomyocyte hypertrophy. Proc Natl Acad Sci USA 107(35):15467–15472
• Shimazu T, Hirschey MD, Newman J, He W, Shirakawa K, Le Moan N et al (2013) Suppression of oxidative stress by beta-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science 339(6116):211–214. This paper suggests that intracellular metabolites can directly regulate HDAC enzyme activity, suggesting this as a novel mechanistic link between metabolism and epigenetics
Ferris GM, Clark JB (1971) Nicotinamide nucleotide synthesis in regenerating rat liver. Biochem J 121(4):655–662
Garcia-Rodriguez JL, Barbier-Torres L, Fernandez-Alvarez S, Juan VG, Monte MJ, Halilbasic E et al (2013) SIRT1 controls liver regeneration by regulating BA metabolism through FXR and mTOR signaling. Hepatology 59:1972–1983
• Jin J, Iakova P, Jiang Y, Medrano EE, Timchenko NA (2011) The reduction of SIRT1 in livers of old mice leads to impaired body homeostasis and to inhibition of liver proliferation. Hepatology 54(3):989–998. These papers investigate the regenerative function of Sirt1
Cihak A, Seifertova M, Vesely J, Sorm F (1972) Metabolic alterations of liver regeneration. 8. Enhanced synthesis of DNA in the liver of 5-azacytidine-treated rats subjected to partial hepatectomy. Int J Cancer 10(1):20–27
Li D, Fan J, Li Z, Xu C (2014) DNA methylation dynamics in the rat EGF gene promoter after partial hepatectomy. Genet Mol Biol 37(2):439–443
Guo G, Xia J, Bao J, Sun HQ, Shi YJ, Bu H (2012) Expression of SOCS3 throughout liver regeneration is not regulated by DNA methylation. Hepatobiliary Pancreat Dis Int 11(4):401–406
Park ES, Park YK, Shin CY, Park SH, Ahn SH, Kim DH et al (2013) Hepatitis B virus inhibits liver regeneration via epigenetic regulation of urokinase-type plasminogen activator. Hepatology 58(2):762–776
Chen L, Zeng Y, Yang H, Lee TD, French SW, Corrales FJ et al (2004) Impaired liver regeneration in mice lacking methionine adenosyltransferase 1A. FASEB J 18(7):914–916
Lu SC, Alvarez L, Huang ZZ, Chen L, An W, Corrales FJ et al (2001) Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc Natl Acad Sci USA 98(10):5560–5565
Shih AH, Abdel-Wahab O, Patel JP, Levine RL (2012) The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer 12(9):599–612
Sedgwick B (2004) Repairing DNA-methylation damage. Nat Rev Mol Cell Biol 5(2):148–157
Song G, Sharma AD, Roll GR, Ng R, Lee AY, Blelloch RH et al (2010) MicroRNAs control hepatocyte proliferation during liver regeneration. Hepatology 51(5):1735–1743
Castro RE, Ferreira DM, Zhang X, Borralho PM, Sarver AL, Zeng Y et al (2010) Identification of microRNAs during rat liver regeneration after partial hepatectomy and modulation by ursodeoxycholic acid. Am J Physiol Gastrointest Liver Physiol 299(4):G887–G897
Raschzok N, Werner W, Sallmon H, Billecke N, Dame C, Neuhaus P et al (2011) Temporal expression profiles indicate a primary function for microRNA during the peak of DNA replication after rat partial hepatectomy. Am J Physiol Regul Integr Comp Physiol 300(6):R1363–R1372
Shu J, Kren BT, Xia Z, Wong PY, Li L, Hanse EA et al (2011) Genomewide microRNA down-regulation as a negative feedback mechanism in the early phases of liver regeneration. Hepatology 54(2):609–619
Raschzok N, Sallmon H, Dame C, Sauer IM (2012) Liver regeneration after partial hepatectomy: inconsistent results of expression screenings for human, mouse, and rat microRNAs. Am J Physiol Gastrointest Liver Physiol 302(4):G470–G471
Zhou J, Ju W, Wang D, Wu L, Zhu X, Guo Z et al (2012) Down-regulation of microRNA-26a promotes mouse hepatocyte proliferation during liver regeneration. PLoS ONE 7(4):e33577
Cirera-Salinas D, Pauta M, Allen RM, Salerno AG, Ramirez CM, Chamorro-Jorganes A et al (2012) Mir-33 regulates cell proliferation and cell cycle progression. Cell Cycle 11(5):922–933
Chen H, Sun Y, Dong R, Yang S, Pan C, Xiang D et al (2011) Mir-34a is upregulated during liver regeneration in rats and is associated with the suppression of hepatocyte proliferation. PLoS ONE 6(5):e20238
Pan C, Chen H, Wang L, Yang S, Fu H, Zheng Y et al (2012) Down-regulation of MiR-127 facilitates hepatocyte proliferation during rat liver regeneration. PLoS ONE 7(6):e39151
• Ng R, Song G, Roll GR, Frandsen NM, Willenbring H (2012) A microRNA-21 surge facilitates rapid cyclin D1 translation and cell cycle progression in mouse liver regeneration. J Clin Invest 122(3):1097–1108. This paper establishes the essential regenerative function of Mir-21
Chen Y, Verfaillie CM (2014) MicroRNAs: the fine modulators of liver development and function. Liver Int 34(7):976–990
Huang J, Schriefer AE, Yang W, Cliften PF, Rudnick DA (2014) Identification of an epigenetic signature of early mouse liver regeneration that is disrupted by Zn-HDAC inhibition. Epigenetics 9:1521–1531
Lee WM (1993) Acute liver failure. N Engl J Med 329(25):1862–1872
Lee WM (2003) Acute liver failure in the United States. Semin Liver Dis 23(3):217–226
Lee WM, Squires RH Jr, Nyberg SL, Doo E, Hoofnagle JH (2008) Acute liver failure: summary of a workshop. Hepatology 47(4):1401–1415
Vetelainen R, van Vliet A, Gouma DJ, van Gulik TM (2007) Steatosis as a risk factor in liver surgery. Ann Surg 245(1):20–30
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection on Stem Cells and Regeneration.
Rights and permissions
About this article
Cite this article
Huang, J., Rudnick, D.A. Elucidating Metabolic and Epigenetic Mechanisms that Regulate Liver Regeneration. Curr Pathobiol Rep 3, 89–98 (2015). https://doi.org/10.1007/s40139-015-0065-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40139-015-0065-3