Abstract
Introduction
Unbiased metagenomic next-generation sequencing (mNGS) has been used for infection diagnosis. In this study, we explored the clinical diagnosis value of mNGS for pulmonary complications after allogeneic hematopoietic stem cell transplantation (allo-HSCT).
Methods
From August 2019 to June 2021, a prospective study was performed to comparatively analyze the pathogenic results of mNGS and conventional tests for bronchoalveolar lavage fluid (BALF) from 134 cases involving 101 patients with pulmonary complications after allo-HSCT.
Results
More pathogens were identified by mNGS than with conventional tests (226 vs 120). For bacteria, the diagnostic sensitivity (P = 0.144) and specificity (P = 0.687) were similar between the two methods. For fungus except Pneumocystis jirovecii (PJ), conventional tests had a significantly higher sensitivity (P = 0.013) with a similarly high specificity (P = 0.109). The sensitivities for bacteria and fungi could be increased with the combination of the two methods. As for PJ, both the sensitivity (100%) and specificity (99.12%) of mNGS were very high. For viruses, the sensitivity of mNGS was significantly higher (P = 0.021) and the negative predictive value (NPV) was 95.74% (84.27–99.26%). Pulmonary infection complications accounted for 90.30% and bacterium was the most common pathogen whether in single infection (63.43%) or mixed infection (81.08%). The 6-month overall survival (OS) of 88.89% in the early group (mNGS ≤ 7 days) was significantly higher than that of 65.52% (HR 0.287, 95% CI 0.101–0.819, P = 0.006) in the late group (mNGS > 7 days).
Conclusions
mNGS for BALF could facilitate accurate and fast diagnosis for pulmonary complications. Early mNGS could improve the prognosis of patients with pulmonary complications after allo-HSCT.
Trial Registration
ClinicalTrials.gov identifier, NCT 04051372.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Why carry out this study? | |
The incidence and mortality of pulmonary complications after allo-HSCT are high, and accurate and fast pathogen diagnosis of pulmonary complications is usually challenging. | |
We aimed to evaluate the diagnostic capacity of mNGS for patients with pulmonary complications after allo-HSCT in the real world. | |
What was learned from the study? | |
mNGS methods have different performance levels for different pathogens in BALF and bacterium is the most common pathogen in lung infection after allo-HSCT. | |
Early mNGS (mNGS ≤ 7 days) for BALF could significantly improve the survival of patients with pulmonary complications after allo-HSCT. |
Introduction
Pulmonary complications are among the most common complications after allogeneic hematopoietic stem cell transplantation (allo-HSCT) with higher incidences of 40–60%, of which 30% died [1]. Pulmonary infection accounts for the majority of pulmonary complications. The diagnosis of lung infection mainly relies on conventional culture of microorganisms, which is time-consuming and insensitive [2]. In clinical practice, accurate and fast diagnosis of pulmonary complications post-transplantation is critical to improve the survival of patients with pulmonary complications, but it is usually challenging.
In recent years, application of bronchoalveolar lavage (BAL) for diagnosis of pulmonary complications has slightly improved the aforementioned dilemma [3]. However, the sensitivity of conventional tests such as culture for bronchoalveolar lavage fluid (BALF) could not be increased significantly [4]. Unbiased metagenomic next-generation sequencing (mNGS) has been used for infection diagnosis, especially for detecting rare or newly emergent pathogens [5], and performs better than traditional methods [6, 7] including diagnosis for pulmonary infection with BALF [8]. Currently most of the studies about mNGS for pulmonary infections focused on patients with normal immunity or critical respiratory failure, and there was little discussion about pulmonary complications following allo-HSCT.
To evaluate the diagnostic capacity of mNGS for patients with pulmonary complications after allo-HSCT, a prospective clinical study was performed in our center. This clinical study was registered at clinicaltrials.org (NCT 04051372). To the best of our knowledge, this is the first report to evaluate the diagnostic value of mNGS for patients after allo-HSCT.
Methods
Patients
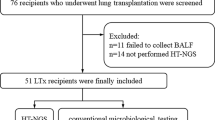
All patients enrolled in the study were admitted to our center because of pulmonary complications after allo-HSCT from August 2019 to June 2021. The following cases were included: (1) patients who had a medical history of allo-HSCT and developed new-onset abnormalities of chest radiography; (2) patients who proceeded with bronchoscopy and whose BALF was obtained. The following cases were excluded: (1) patients who failed to undergo bronchoscopy as a result of severe dyspnea and platelet counts below 30 × 109/L; (2) BALF samples failed to pass quality control for mNGS; (3) patients with incomplete clinical and laboratory data; (4) patients who were lost to follow-up. An independent pulmonary complication was considered within one patient with an interval of at least 4 weeks between two separate pulmonary complications. The study was conducted in accordance with the principles of the Declaration of Helsinki and approved by the ethics committee of Shanghai General Hospital (2018KY270). Informed consents were obtained before BAL.
Pathogens Identified by Conventional Tests
-
1.
Bacteria: The number of bacteria above 104 CFU/ mL was considered as a positive criterion for culture with BALF and sputum [9].
-
2.
Fungi: Fungi found in cultures and smears from BALF and sputum were considered as a positive result. Galactomannan (GM) antigen detection in BALF and peripheral blood was positive for pulmonary aspergillosis, while beta-d-glucan was positive for fungus, including Pneumocystis jirovecii (PJ) [10]. PJ was confirmed by trophozoite and carinii cyst of BALF samples [11].
-
3.
Viruses: Cytomegalovirus (CMV) and Epstein-Barr virus (EBV) were defined by polymerase chain reaction (PCR), and 500 IU/ml was defined as the cutoff value for CMV [12, 13]. Other viruses such as influenza virus were detected with enzyme‐linked immunosorbent assays, and multiplex PCR panels were not used to detect other respiratory viruses.
-
4.
Tuberculosis: Tuberculosis was defined by PCR (at any level) with BALF samples. Acid-fast bacilli and culture for TB can also be used to make a definitive diagnosis. The conventional tests were performed on the same day as BAL.
Pathogens of BALF Identified by mNGS
Approximately 5 ml BALF sample was immediately sent for mNGS analysis. DNA libraries were established after DNA extraction as reference [14]. High-quality sequencing data were generated by removing low-quality reads, followed by computational subtraction of human host sequences mapped to the human reference genome (hg19) using Burrows-Wheeler Alignment [15] and were classified by simultaneously aligning to four Microbial Genome Databases, consisting of bacteria, fungi, viruses, and parasites. The classification reference databases were downloaded from NCBI (ftp://ftp.ncbi.nlm.nih.gov/genomes/). RefSeq contains 4945 whole genome sequences of viral taxa, 6350 bacterial genomes or scaffolds, 1064 fungi related to human infection, and 234 parasites associated with human diseases.
Criteria for a Positive mNGS Result
As a result of the lack of a standard method for analyzing the results of mNGS for pathogens, especially in immunodeficient patients, we used the following criteria in this study [6, 16,17,18]. To standardize the results, the specifically mapped read number (SMRN) of each microbial taxonomy was normalized to SMRN per 20 million (M) of total sequencing reads (SDSMRN, standardized SMRN). The suspected background microorganisms and normal flora of the oral cavity, respiratory tract, or skin were removed from the microbial list after a large number of literature searches.
For different types of microbes, the thresholds were set as follows:
-
1.
Bacterium/mycoplasma/chlamydia/fungus: SDSMRN ≥ 3
-
2.
Virus: SDSMRN ≥ 3
-
3.
Parasite: SDSMRN ≥ 100
-
4.
Mycobacterium tuberculosis: SDSMRN ≥ 1
Comprehensive Clinical Diagnosis
Two hematologists independently reviewed the medical records of all cases including clinical manifestations, chest radiology, laboratory tests, microbiological tests (conventional tests and mNGS), and treatment response. The causative pathogens of pulmonary infection were determined according to the comprehensive clinical characteristics and available literature. When two hematologists could not reach a consensus on the diagnosis for a patient, a respiratory physician would be invited for an in-depth discussion to solve the disagreement according to related guidelines [9, 10, 19,20,21,22,23].
Statistical Analysis
According to the collected data, the chi-squared test and t test were applied to the proportions and measurement data compared, separately. 2 × 2 contingency tables were established to calculate sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). Wilson’s method was used to determine the 95% confidence interval. Kaplan–Meier method was used to compare the 6-month overall survival and no-relapse mortality after pulmonary complications. P values less than 0.05 were considered to indicate significant differences. All tests were two-sided. Statistical analyses were conducted by SPSS Version 22.0 and figures were prepared with GraphPad Prism 8.0.
Results
Patient Characteristics
A total of 134 cases involving 101 patients with pulmonary complications were enrolled into the study (Fig. 1). Out of the 101 patients, 75 underwent BAL only once, while the remaining 26 underwent BAL more than once (21 twice, 4 three times, and 1 five times). Baseline characteristics of all patients are shown in Table 1.
Detection of Pathogens
A total of 266 strains of pathogens (Table S1 A–C) were identified by mNGS, including 87 (87/266, 32.71%) of bacteria, 33 (33/266,12.41%) of fungi, and 146 (146/266, 54.89%) of viruses. The most frequent species of bacteria were Pseudomonas aeruginosa (n = 17, 6.39%) and Klebsiella pneumoniae (n = 8, 3.01%) (Fig. 2a). The most frequent species of fungi were PJ (n = 22, 8.27%), followed by Candida (n = 6, 2.25%). Aspergillus was only found in two cases (Fig. 2b). Among the viruses, the most common were human beta-herpesvirus 5 (n = 50, 18.79%) and human gamma-herpesvirus 4 (n = 35, 13.16%) (Fig. 2c). At the same time, a total 120 strains of pathogens were identified by conventional tests, including bacteria (n = 48, 40%), fungi (n = 19, 15.83%), PJ (n = 2, 1.67%), and viruses (n = 51, 42.5%) (Fig. 2d).
Comparison of mNGS and Conventional Tests
The comparison of pathogens detected by mNGS and conventional tests is shown in Fig. 2e and Table S2. The detection frequencies for bacteria between mNGS and conventional tests were similar [(87/266, 32.71%) vs (48/120, 39.67%), P = 0.164], but the detection rate for Gram positive bacteria by mNGS was significantly higher than by conventional tests [(35/87, 40.23%) vs (11/48, 22.92%), P = 0.042]. The detection rate for viruses by mNGS was higher than by conventional tests [(146/266, 54.89%) vs (51/120, 42.5%), P = 0.024], whereas the positive rate for fungi except for PJ was significantly lower by mNGS than by conventional tests [11(11/266, 4.14%) vs (19/120, 15.83%), P = 0.000]. The detection rate for mold was also lower than that of Saccharomyces by mNGS [(3/11, 27.27%) vs (8/11, 72.73%, P = 0.088]. It should be noted that for PJ, the detection rate of 8.27% (22/266) by mNGS was significantly higher than that of 1.67% (2/120) by conventional tests (P = 0.013). For viruses, more respiratory viruses (except CMV and EBV) were detected by mNGS. Among the pathogens detected by mNGS (n = 266), 99 strains were considered as “false positive” results according to clinical diagnosis, which virus accounted for 89.90% (89/99) (Table S3); 30 strains of pathogens were considered “false negative” by mNGS, of which Gram negative bacteria and fungi (except for PJ) accounted for 43.33% (13/30) and 36.36% (12/30), respectively (Table S4).
Concordance of Results Between mNGS and Conventional Tests
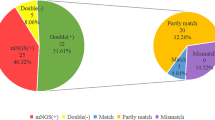
Out of the 134 cases, 32 (23.88%) had concordant results between mNGS and conventional tests, while 102 (76.12%) cases did not. Out of the 32 cases with concordant results, 22 (68.75%) were both positive and completely matched, and 10 (31.25%) were both negative. Among the 22 completely matched cases, one CMV-positive case was considered as contamination from viremia, while the other 21 cases were accurately diagnosed and the diagnostic coincidence rate was 95.45% (21/22) (Fig. S1). For the 42 partly matched cases, the false positive and false negative results of pathogens are shown in Table S5. In addition, the details of concordance results of the nine mismatched cases and all the cases are shown in Tables S6 and S7, respectively.
Diagnostic Performance of mNGS and Conventional Tests
The sensitivities and specificities of conventional tests, mNGS, and the combination of the two methods for diagnosis of pulmonary complications are shown in Table 2. For bacteria, there were no significant differences of the sensitivity (P = 0.144) and specificity (P = 0.687) between mNGS and conventional tests, while the sensitivity could be increased (to 84.15%), but the specificity could not when the two methods were combined. Some potential bacterial pathogens were detected by mNGS such as Trophery mawhipplei, Mycobacterium tuberculosis, Mycobacterium abscessus, and Legionella pneumophil, which were not detected by conventional tests. For fungi (excluding PJ), conventional tests had a higher sensitivity than mNGS (P = 0.013), while the two methods had similar high specificities (P = 0.109). The sensitivity for fungi could be increased to 96% (95% CI 77.68–99.79%) with the combination of the two methods. As for PJ, both the sensitivity (100%) and specificity (99.12%) of mNGS were very high, while the sensitivity (9.52%) of conventional tests was very low, although the specificity (100%) was very high. For viruses, the sensitivity of mNGS was significantly higher (P = 0.021) and the specificity was significantly lower (P = 0.003) than with conventional tests. It should be noted that the NPV was 95.74% (95% CI 84.27–99.26%) for viruses by mNGS.
Diagnosis and Classification of Pulmonary Complications
According to clinical manifestations, radiological characteristics, microorganism results, and response to treatment, 90.30% (121/134) of cases were diagnosed with infectious complications and 9.70% (13/134) with non-infectious complications. A single pathogen infection was the most common type with a frequency of 63.43% (85/134), followed by mixed infection (36/134, 26.87%). Among all the pathogens of pulmonary infectious complications including single and mixed infections, the most common pathogen was bacterium accounting for 59.87%, followed by PJ for 21.39%, fungi for 21.22%, and virus for 18.92%. The diagnosis distributions of 134 cases are shown in Fig. S2.
Patient Management and Outcomes
According to clinical diagnosis, treatment response, and mNGS results, the antibiotic coverage was narrowed for 32 cases, broadened for 47 cases, while the original antibiotic coverage was maintained for 15 cases (Table S8). Among all the patients with follow-up for at least 6 months since the last diagnosis of pulmonary complications, 18 patients died, of which 11 died from pulmonary complications (Table S9). The overall mortality was 17.82% (18/101) and the mortality of pulmonary complications was 10.89% (11/101). Patients with pulmonary complications were divided into two groups according to the time of BAL for mNGS from the initiation of pulmonary complication: early group (mNGS ≤ 7 days) and late group (mNGS > 7 days), containing 72 and 29 patients, respectively. The patients’ characteristics were similar between the two groups (P > 0.05) (Table S10). The 6-month overall survival (OS) of 88.89% in the early group was significantly higher than that of 65.52% (HR 0.287, 95% CI 0.101–0.819, P = 0.006) in the late group, while no-relapse mortality (NRM) of 9.72% was significantly lower than that of 27.59% (HR 0.313, 95% CI 0.099–0.986, P = 0.019), which indicated that early BAL and mNGS could improve the prognosis of patients with pulmonary complications after allo-HSCT (Fig. 3).
Discussion
In this study, mNGS performance better with detection of more pathogens (266 vs 120) and higher sensitivity (80.99% vs 59.50%, P < 0.001) than conventional tests and performed differently for different pathogens [8, 24]. The sensitivity of mNGS for bacteria was similar to that of conventional tests (65.85% vs 52.44%, P = 0.144); whereas for fungi (except for PJ), it was significantly lower (36% vs 80%, P = 0.013). These results were opposite to those from Miao’s study [7] because of only 29.16% (149/511) of BALF samples in the latter [24]. The sensitivity for bacteria could be increased to 84.15% with the combination of the two methods. These results suggested that mNGS might be an important supplement to conventional tests to detect some potential pathogens such as Mycobacterium genus [7, 25] and increase the detection sensitivity for bacteria.
For fungi except for PJ, the sensitivity of mNGS was significantly lower than that of conventional tests [26].The major reason might be that the lower sensitivity for Aspergillus genera by mNGS may due to having hard-to-break cell walls [27]. For PJ, we found that mNGS achieved a high sensitivity of 100% and a specificity of 99% in our study [28]. Conventional tests had a significantly lower sensitivity of 9.52% for PJ in the present study because only a morphological method was used for detecting trophozoite and/or carinii cysts. Trichosporon and Penicillium genera were also detected by mNGS in the present study, which was important for patients with pulmonary complications after allo-HSCT because more and more rare fungi such as Trichosporon pneumonia were reported [24, 29]. These results suggested that a diagnosis of PJ pneumonia should be considered when a positive result for PJ by mNGS was obtained, while a diagnosis for Aspergillus pneumonia should not be excluded when a negative result for Aspergillus is obtained by mNGS.
In terms of viruses, mNGS had a higher sensitivity (93.94% vs 69.70%, P = 0.021), but lower specificity (44.55% vs 79.21%, P = 0.003) than with conventional tests. In the present study, except for CMV and EBV, nearly 60% of other potential pathogenic viruses were detected with mNGS, while conventional tests only covered CMV and EBV, which might lower the sensitivity of conventional tests for viruses. The NPV for viruses by mNGS was above 95%, which suggested that a virus pneumonia could be nearly excluded if a negative result was obtained by mNGS [18].
Pulmonary infection complications accounted for 90.30%, while non-infectious complications accounted for 9.7%. The incidences of various pathogens in pulmonary infection complications are different at different times after allo-HSCT [1]. Although the median time in our study from allo-HSCT to pulmonary complications was 154.5 days, the bacteria were the most common pathogens in both single and mixed pulmonary infection. However, we found that the detection rate of Gram positive bacteria by mNGS was higher than that of conventional tests (40.23% vs 22.92%, P = 0.042). This was probably because a number of Gram positive bacteria have been missed in the past, with about half of the cases (43/101, 42.57%) developing graft-versus-host disease and 85.15% of patients receiving myeloablative conditioning regimen, which could make them susceptible to bacteria [30].
The mortality of pulmonary complications was 8.21% (11 /101) in our study, which was lower than that of 18–52.46% in previous reports [31, 32]. This might be due to the benefit of early accurate diagnosis and discerning the pathogens by mNGS in combination with conventional tests for BALF. Notably, early mNGS for BALF (≤ 7 days) significantly decreases the NRM compared with the late mNGS (> 7 days) (9.72% vs 27.59%, P = 0.019), which improved the OS of patients with pulmonary complications (88.89% vs 65.52%, P = 0.006). Our study suggested that early mNGS for BALF would be critical to early accurate diagnosis and would improve the survival of patients with pulmonary complications after allo-HSCT.
Finally, our study had limitations. It was a single-center retrospective study and the sample size of pulmonary complications after HSCT was limited. The positive result of mNGS could not indicate whether the pathogen was alive, colonized, or pathogenic, which needs to be further analyzed combined with clinical data.
Conclusions
Our findings suggest that mNGS could facilitate the fast and accurate diagnosis of pulmonary complications. At the same time, early BAL and mNGS for pathogens could improve the survival of patients with pulmonary complications after HSCT. As a result of the limitations of this single-center study and its sample size, multicenter prospective studies with a large sample size are required.
References
Sirithanakul K, Salloum A, Klein JL, et al. Pulmonary complications following hematopoietic stem cell transplantation: diagnostic approaches. Am J Hematol. 2005;80(2):137–46.
Maschmeyer G, Carratalà J, Buchheidt D, et al. Diagnosis and antimicrobial therapy of lung infiltrates in febrile neutropenic patients (allogeneic SCT excluded): updated guidelines of the Infectious Diseases Working Party (AGIHO) of the German Society of Hematology and Medical Oncology (DGHO). Ann Oncol. 2015;26(1):21–33.
Shannon VR, Andersson BS, Lei X, et al. Utility of early versus late fiberoptic bronchoscopy in the evaluation of new pulmonary infiltrates following hematopoietic stem cell transplantation. Bone Marrow Transplant. 2010;45(4):647–55.
Hummel M, Rudert S, Hof H, et al. Diagnostic yield of bronchoscopy with bronchoalveolar lavage in febrile patients with hematologic malignancies and pulmonary infiltrates. Ann Hematol. 2008;87(4):291–7.
Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–74.
Qian YY, Wang HY, Zhou Y, et al. Improving pulmonary infection diagnosis with metagenomic next generation sequencing. Front Cell Infect Microbiol. 2020;10: 567615.
Miao Q, Ma Y, Wang Q, et al. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin Infect Dis. 2018;67(suppl_2):S231–S40.
Langelier C, Zinter MS, Kalantar K, et al. Metagenomic sequencing detects respiratory pathogens in hematopoietic cellular transplant patients. Am J Respir Crit Care Med. 2018;197(4):524–8.
Miller JM, Binnicker MJ, Campbell S, et al. A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2018 update by the Infectious Diseases Society of America and the American Society for Microbiology. Clin Infect Dis. 2018;67(6):e1–94.
Donnelly JP, Chen SC, Kauffman CA, et al. Revision and update of the consensus definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium. Clin Infect Dis. 2020;71(6):1367–76.
Tissot F, Agrawal S, Pagano L, et al. ECIL-6 guidelines for the treatment of invasive candidiasis, aspergillosis and mucormycosis in leukemia and hematopoietic stem cell transplant patients. Haematologica. 2017;102(3):433–44.
Ljungman P, Boeckh M, Hirsch HH, et al. Definitions of cytomegalovirus infection and disease in transplant patients for use in clinical trials. Clin Infect Dis. 2017;64(1):87–91.
Kotton CN, Kumar D, Caliendo AM, et al. The third international consensus guidelines on the management of cytomegalovirus in solid-organ transplantation. Transplantation. 2018;102(6):900–31.
Jeon YJ, Zhou Y, Li Y, et al. The feasibility study of non-invasive fetal trisomy 18 and 21 detection with semiconductor sequencing platform. PLoS One. 2014;9(10): e110240.
Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–60.
Benamu E, Gajurel K, Anderson JN, et al. Plasma microbial cell-free DNA next generation sequencing in the diagnosis and management of febrile neutropenia. Clin Infect Dis. 2022;74(9):1659–68.
Zhang NN, Huang X, Feng HY, et al. Circulating and pulmonary T-cell populations driving the immune response in non-HIV immunocompromised patients with Pneumocystis jirovecii pneumonia. Int J Med Sci. 2019;16(9):1221–30.
Wang S, Ai J, Cui P, et al. Diagnostic value and clinical application of next-generation sequencing for infections in immunosuppressed patients with corticosteroid therapy. Ann Transl Med. 2020;8(5):227.
Pappas PG, Kauffman CA, Andes D, et al. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis. 2009;48(5):503–35.
Ljungman P, de la Camara R, Robin C, et al. Guidelines for the management of cytomegalovirus infection in patients with haematological malignancies and after stem cell transplantation from the 2017 European Conference on Infections in Leukaemia (ECIL 7). Lancet Infect Dis. 2019;19(8):e260–e72.
Boeckh M, Stevens-Ayers T, Travi G, et al. Cytomegalovirus (CMV) DNA quantitation in bronchoalveolar lavage fluid from hematopoietic stem cell transplant recipients with CMV pneumonia. J Infect Dis. 2017;215(10):1514–22.
Alanio A, Hauser PM, Lagrou K, et al. ECIL guidelines for the diagnosis of Pneumocystis jirovecii pneumonia in patients with haematological malignancies and stem cell transplant recipients. J Antimicrob Chemother. 2016;71(9):2386–96.
Green ML. Viral pneumonia in patients with hematopoietic cell transplantation and hematologic malignancies. Clin Chest Med. 2017;38(2):295–305.
Guo LN, Yu SY, Hsueh PR, et al. Invasive infections due to trichosporon: species distribution, genotyping, and antifungal susceptibilities from a multicenter study in China. J Clin Microbiol. 2019;57(2):e01505–18.
Zhou X, Wu H, Ruan Q, et al. Clinical evaluation of diagnosis efficacy of active mycobacterium tuberculosis complex infection via metagenomic next-generation sequencing of direct clinical samples. Front Cell Infect Microbiol. 2019;9:351.
Xie G, Zhao B, Wang X, et al. Exploring the clinical utility of metagenomic next-generation sequencing in the diagnosis of pulmonary infection. Infect Dis Ther. 2021;10(3):1419–35.
Gow NAR, Latge JP, Munro CA. The fungal cell wall: structure, biosynthesis, and function. Microbiol Spectr. 2017;5(3). https://doi.org/10.1128/microbiolspec.funk-0035-2016.
Jiang J, Bai L, Yang W, et al. Metagenomic next-generation sequencing for the diagnosis of Pneumocystis jirovecii pneumonia in non-HIV-infected patients: a retrospective study. Infect Dis Ther. 2021;10(3):1733–45.
de Almeida Júnior JN, Hennequin C. Invasive trichosporon infection: a systematic review on a re-emerging fungal pathogen. Front Microbiol. 2016;7:1629.
Bondeelle L, Bergeron A. Managing pulmonary complications in allogeneic hematopoietic stem cell transplantation. Expert Rev Respir Med. 2019;13(1):105–19.
Lucena CM, Torres A, Rovira M, et al. Pulmonary complications in hematopoietic SCT: a prospective study. Bone Marrow Transplant. 2014;49(10):1293–9.
Aguilar-Guisado M, Jiménez-Jambrina M, Espigado I, et al. Pneumonia in allogeneic stem cell transplantation recipients: a multicenter prospective study. Clin Transplant. 2011;25(6):E629–38.
Acknowledgements
We thank the participants in the study.
Author Contribution
Jun Yang, Yu Cai, Liping Wan, Chongmei Huang, Xiaowei Xu, and Jiahua Niu conceptualized the study, secured research funding, contributed to study design, and reviewed the manuscript. Zaihong Shen, Ying Wang and Aihua Bao drafted the initial manuscript, contributed to the analysis, and revised the manuscript. Xinxin Xia, Chang Shen, Yu Wei, Huiying Qiu, Kun Zhou conducted data analysis and reviewed the manuscript. Min Zhang, Yin Tong and Xianmin Song provided methodological and analytical input and reviewed the manuscript. All authors approved the manuscript.
Funding
This research was funded by Three-year development project from Shanghai Shen Kang Hospital Development Center (SHDC2020CR1012B for Xianmin Song); Shanghai Science and Technology Commission Western Medicine Guidance Project (19411970000 for Yin Tong); Clinical Research Special Clinical Research Innovation Plan of Shanghai General Hospital (CTCCR-2019D02 for JiahuaNiu; CTCCR-2019B03 for Chongmei Huang). The rapid service fee was funded by the authors.
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethical Approval
The study was conducted in accordance with the principles of the Declaration of Helsinki and approved by the ethics committee of Shanghai General Hospital (2018KY270). Informed consents were obtained before BAL.
Conflict of Interest
Zaihong Shen, Ying Wang, Aihua Bao, Jun Yang, Xi Sun, Yu Cai, Liping Wan, Chongmei Huang, Xiaowei Xu, Jiahua Niu, Xinxin Xia, Chang Shen, Yu Wei, Huiying Qiu, Kun Zhou, Min Zhang, Yin Tong, Xianmin Song. All have nothing to disclose.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Shen, Z., Wang, Y., Bao, A. et al. Metagenomic Next-Generation Sequencing for Pathogens in Bronchoalveolar Lavage Fluid Improves the Survival of Patients with Pulmonary Complications After Allogeneic Hematopoietic Stem Cell Transplantation. Infect Dis Ther 12, 2103–2115 (2023). https://doi.org/10.1007/s40121-023-00850-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40121-023-00850-w