Abstract
Introduction
Cytomegalovirus (CMV) predisposes to several clinical complications and is a major cause of morbidity and mortality in immunocompromised patients, including patients with hematological malignancies (HM). The present study was carried out to determine the distribution of CMV glycoprotein B, N, and O (gB, gN, and gO) genotypes and their potential effect on its viral load and on clinical outcomes in a cohort of Tunisian non-hematopoietic stem cell transplant (HSCT) patients with HM undergoing chemotherapy.
Methods
CMV viral load was evaluated by real-time quantitative PCR. The gB, gN, and gO genotypes of the CMV strains were analyzed by multiplex nested PCR and sequencing.
Results
This prospective study involved 60 clinical isolates obtained from 60 non-HSCT patients with HM undergoing chemotherapy. Mixed CMV gB, gN, and gO genotypes were the predominant glycoprotein genotypes in 31%, 41.4%, and 46.4% of patients, respectively. Mixed gB genotypes were associated with higher initial levels of CMV load (p = 0.001), increased rate of fever (0.025), and co-infection with other herpesviruses (HHVs) (p = 0.024) more frequently than in single gB genotype. Mixed gN genotypes were more associated with severe lymphopenia (ALC < 500/µL) (p = 0.01) and increased risk of death (p = 0.042) than single gN genotype. Single gO2b genotype had also a more unfavorable outcome (p = 0.009) than the other single gO genotype. Mixed gO genotypes were associated with female gender (p = 0.015), acute leukemia disease (p = 0.036), initial high level of CMV viral load (at least 1000 copies/mL) (p = 0.029), skin rash (p = 0.01) more frequently than in single gO genotype. The gO1a/gN3b linkage was associated with an increased initial viral load (p = 0.012).
Conclusion
Infection with mixed CMV genotypes was common and multiple gB, gN, and gO genotypes were associated with clinical manifestation and higher viral load.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Why carry out this study? |
There are limited data on CMV genotypes among non-hematopoietic stem cell transplant (HSCT) patients with hematological malignancies (HM) undergoing chemotherapy. |
This study aimed to determine the distribution of CMV gB, N, and O genotypes and to explore their potential effect on its viral load and on clinical outcomes. |
What was learned from the study? |
Our study indicated that mixed CMV gB, gN, and gO genotypes were common. |
Mixed glycoprotein genotypes were more frequently associated with clinical manifestation and higher viral load. |
Digital Features
This article is published with digital features, including a summary slide, to facilitate understanding of the article. To view digital features for this article go to https://doi.org/10.6084/m9.figshare.14560542.
Introduction
Patients at risk of severe cytomegalovirus (CMV) infection include preterm neonates, patients with congenital or acquired immunodeficiency such as HIV, those undergoing chemotherapy or radiotherapy, and solid organ or hematopoietic stem cell transplant (HSCT) patients. CMV infections and subsequent diseases are increasingly recognized in non-transplant hematologic malignancies settings, especially in leukemia and lymphoma receiving chemotherapy. In these patients, CMV may cause either mild or even asymptomatic clinical manifestations as well as life-threatening disease [1,2,3]. However, little data are available about the impact of CMV virulence factors on infection outcome [4]. The virulence among different CMV strains may be an important factor in the occurrence and severity of CMV-associated diseases [5, 6], because of the genetic variation in functionally important genes such as glycoproteins gB, gN, and gO that are involved in host cell penetration, tissue tropism, or replication [6,7,8,9,10,11,12,13]. Five gB genotype (gB1, gB2, gB3, gB4, and gB5) have been identified on the basis of sequence variation in the UL55 gene encoding gpUL55 (gB) [14, 15]. The gpUL73 (gN) encoded by UL73 gene has four main genomic variants gN1, gN2, gN3 and gN4, with gN3 subdivided into gN3a and gN3b while the gN4 genotype has three subgroups gN4a, gN4b, and gN4c [16, 17]. Another CMV surface glycoprotein, gpUL74 (gO), encoded by UL74 gene has at least eight genetic variants, including five major genotypes (gO1, gO2, gO3, gO4, and gO5) with minor subtypes (gO1a, gO1b, gO1c, gO2a, gO2b) [9]. In immunocompromised patients, mixed CMV infection has been shown to be associated with elevated viral load [5]. However, there is no full understanding of the pathogenic mechanisms underlying this association. The relationship between CMV genetic variants and serious complications of CMV infection can depend on the additive or synergistic activity of at least two CMV glycoproteins. No evidence of genetic linkage between gN/gB genotypes has been observed previously [17]. On the other hand, there were eight significant linkages between the gN/gO genotypes as follows: gO1a/gN1, gO1b/gN3a, gO1c/gN4c, gO2a/gN3b, gO2b/gN2, gO3/gN4a, gO4/gN4b, and gO5/gN4c from different clinical settings [18, 19]. The impact of these linkages on the severity of CMV infection is not well defined. The genetic linkages between gO and gN could have a potentially functional interaction, which may lead to virus attachment, replication, and spread.
Thus, the current study investigated the distribution of CMV gB, N, and O genotypes and explored the possible relationship between different genotypes and clinical characteristics in Tunisian non-HSCT patients with HM.
Methods
Study Population
To screen patients for our study, 95 patients with newly diagnosed acute leukemia (52 acute lymphoblastic leukemia (ALL) (39 B cell ALL and 13 T cell ALL) and 43 acute myeloid leukemia (AML)) and 50 Hodgkin’s and non-Hodgkin’s lymphoma (10 HL and 40 NHL) were prospectively enrolled in the follow-up study during the period from January 2016 to December 2018. All 145 consecutive patients with leukemia/lymphoma were included, regardless of CMV serostatus. Initial treatment with conventional chemotherapy induces initial remission. Consolidation and maintenance therapy have been used as post-remission strategies. Salvage regimens were selected in case of primary refractory or at relapse. In the present study, patients who received allogeneic or autologous HSCT were excluded. During the study period, patients included did not receive CMV antiviral therapy. Most viral infections were self-limited (clear the virus without therapy).
Study Design
The study design involved prospective sampling at the time of diagnosis (before chemotherapy) and in the different phases of chemotherapy (post-induction, post-remission, and post-salvage). Plasma samples were collected from patients at diagnosis, and once every 7–14 days in the same period, but the interval was greater because sampling was not performed during periods in which patients were not hospitalized. For some patients, the number of follow-ups was limited because of early death or loss of follow-up samples. The median follow-up duration was 7.4 months range (1–36) from the start of chemotherapy for patients with acute leukemia and 6.8 months range (2–15) for those with lymphoma. Only those patients with positive CMV infection were included in this study and were evaluated with CMV load testing and genotyping of CMV glycoproteins B, N, and O. These patients included a cohort of 60 non-HSCT patients with active CMV infection.
The study design was approved by the Ethics Committee and Medical Research of the Farhat Hached University Hospital, Sousse, Tunisia (Reference number IRB00008931) and informed consent was obtained from all participants enrolled in the study before starting the research work.
Definitions and Data Collected
Active CMV infection was defined as the detection of CMV DNA (any level of plasma viral load) in plasma specimens. For each patient, CMV recurrence was defined as detectable CMV DNAemia after initially achieving clearance following the initial episode of CMV infection. A viral co-infection was assigned as the detection of DNA of more than one type of herpesvirus (HHV) in the same sample and mixed genotype was defined as the presence of more than one CMV genotype in the same sample.
The hospital records were evaluated and the clinical characteristics and microbiological data were registered for each patient. Detailed information regarding the data collected in the present study is provided in the supplementary material.
CMV Serology
Serum samples from all patients before starting chemotherapy were assessed for anti-CMV IgG antibodies with chemiluminescence assays by the Abbott Architect i2000SR.
Multiplex PCR Detection for Herpesviruses (HHVs)
HHVs DNA were extracted from plasma samples using a DNA extraction kit (QIAamp DNA Mini Kit, QIAGEN, Germany). The detection of HHVs (HSV-1/2, VZV, EBV, CMV, HHV-6A/B, and HHV-7) was performed on plasma samples according to the multiplex PCR method described by Tanaka et al. [20].
Quantitative PCR Assay for CMV DNA
Only those patients with HHVs multiplex results positive for CMV were included in the study and evaluated with CMV viral load in the first positive plasma samples using the artus CMV RG PCR kit (Qiagen, Germany) and the Rotor-Gene Q (RGQ) instrument according to the manufacturer’s recommendations. The lower limit of detection for this assay is 57.1 copies/mL. On the basis of the assay results, patients were categorized as patients with high-level CMV DNAemia (at least 1000 copies/mL) and low/moderate-level DNAemia (less than 1000 copies/mL).
Characterization of CMV gB, gN, and gO Genotypes
The first samples determined to be CMV DNA positive were further examined by genotyping envelope glycoproteins, UL55 (gB), UL73 (gN), and UL74 (gO) by the multiplex nested PCR assays as previously described [11, 21, 22].
The primers are listed in Table S1 in the supplementary material. The primers for the first round of PCR were designed on the conserved regions of UL55 (gB), UL73 (gN), and UL74 (gO) gene. After the first round of PCR, the amplicons were subjected to the next multiplex PCR round resulting in genotype determination using a set of genotype-specific primers. Positive and negative controls were included with each PCR run. Then the different genotypes were easily differentiated by agarose gel electrophoresis.
DNA Sequencing and Sequences Analysis
Randomly selected amplicons for the different gB/gO genotyping results were then confirmed by sequencing. Both forward and reverse DNA strands were sequenced with the same specific primers of the first PCR round using an ABI PRISM Big Dye Terminator Cycle Sequencing Reaction kit (version 3.1) on an ABI PRISM 3100 DNA sequencer (Applied Biosystems, Germany). Part of the gN genotyping results were further confirmed by PCR and sequencing using the gN sequencing primers. Sequencing alignment analysis was conducted using the NCBI BLAST to confirm their viral genotype.
Statistical Analysis
All statistical analyses were performed using SPSS software (version 22.0). The results are expressed as the mean ± standard deviation (SD), and percentage. Categorical variables were compared using the chi-square test or the Fisher’s exact test. All p values were based on a two-tailed test of significance. Statistical significance was defined as a p value less than 0.05.
Results
Patients’ Characteristics
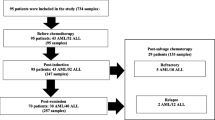
Flowchart of patient enrollment is presented in Fig. 1. The overall CMV IgG seroprevalence was 96.5% (140/145). None of the patients who were negative for CMV IgG developed CMV DNAemia during the follow-up study. The prevalence of CMV DNAemia was 60 out of 145 (41.4%). According to each period, CMV DNAemia was detected in 5.5% (8/145), 11.1% (16/145), 16.4% (18/110), and 33.33% (18/54) before chemotherapy, at post-induction, post-remission, and post-salvage, respectively. Table 1 summarizes the demographic characteristics and clinical data of the enrolled patients. Sixty patients with active CMV infection (46 acute leukemia and 14 lymphomas) were included in the study between 2016 and 2018. The male/female sex ratio was 2.5. The patients were aged 1–69 years (mean 24).
Detection and Quantitative PCR Assay for CMV DNA
Of the 60 patients infected with CMV, 39 (65%) patients were single infected and 21 (35%) were co-infected with other HHVs (EBV, HHV-6, HHV-7); 19 of these with two viruses and 2 with three viruses. None of those patients were positive for HSV-1, HSV-2, and VZV. All patients with active CMV infection were CMV IgG positive before starting chemotherapy.
High level of CMV DNAemia (at least 1000 copies/mL) was developed in half of the patients (30/60). All high-level cases of CMV DNAemia were at post-remission and at post-salvage phase of chemotherapy and not before chemotherapy.
Distribution of gB, gN, and gO Genotypes in CMV-Infected Patients
The distributions of glycoprotein gB, N, and O genotype are presented in Fig. 2. As observed, all the expected genotypes are present for gB, gN, and gO except for the gB5, gN2, and gN4b genotypes, none of which were present in the studied population. Sixty percent of CMV-infected patients (36/60) were found to carry mixed infections with multiple genotypes in at least one genetic locus of gB, gN, and gO. The gB, gN, and gO genotype was successfully determined in 96.6% (58/60), 93.3% (56/60), and 93.3% (56/60) of CMV-infected patients, respectively. Mixed CMV gB, gN, and gO genotypes were detected in 31% (18/58), 41% (23/56), and 46.4% (26/56) of CMV-infected patients, respectively. Among mixed gB genotypes, 72% (13/18) were dual, 22.2% (4/18) were triple, and only one was a quadruple genotype, i.e., 5.5% (1/18). Among the gN mixed genotypes, 87% (20/23) were dual gN genotype and 13% (3/23) triple genotype. Among the mixed gO genotypes, 76.9% (20/26) were dual gO genotypes and 23.1% (6/26) were triple genotypes.
Twelve patients in our cohort developed a second recurrent episode of CMV infection. Between the first and the follow-up samples investigated in the present study, all patients showed periods of undetectable CMV DNAemia. The median time between the initial and the relapse episode was 9.5 months range (2–16). Variations in glycoproteins were observed between initial and recurrent infection. An increase in the complexity of these genetic populations was seen in half of the cases, and new genotypes emerged compared to those found in the initial episode, while the others maintained the same genotypes, found in the initial episode, throughout the follow-up.
To evaluate further the accuracy of the multiplex PCR based, 24 single genotype strains were sequenced. There was no discrepancy between subtyping based on multiplex PCR assays and UL55 (gB), UL73 (gN), and UL74 (gO) sequences (data not shown).
Analysis of Glycoprotein Genotypes and Clinical Characteristics in CMV-Infected Patients
The relationship between CMV (gB) single genotypes and mixed genotypes and clinical characteristics of patients are presented in Table 2. When comparing infections involving mixed gB genotype infections and single genotype infections no statistically significant differences were observed with respect to gender status, age groups (≤ 22 years, > 22 years), underlying disease, status of the disease, recurrence of infection, clinical findings (severe neutropenia, febrile neutropenia, severe lymphopenia, anemia, thrombocytopenia, pneumonia, gastroenteritis, hepatitis, CNS disease, and skin rash), and opportunistic and infectious complications (bacteremia/fungal infections). Table 2 demonstrates that patients infected with mixed gB genotypes developed higher initial levels of CMV loads (p = 0.001), had an increased rate of fever (p = 0.025) ,and were significantly more often infected with other herpes viruses (p = 0.024) than patients with single gB genotype.
In Table 3, when comparing infections involving only single gN genotypes, single genotypes had common characteristics including gender status, age groups (≤ 22 years, > 22 years), underlying disease, status of the disease, recurrence of infection, intensity of initial viral load, clinical findings, and opportunistic and infectious complications.
The presence of mixed gN genotypes was more frequently associated with profound lymphopenia (ALC < 500/µL) (p = 0.01) and increased risk of death (p = 0.042) than with single gN genotype, but this finding was not time-related (Table 3).
Because the number of infected patients with gO1a, gO1b, gO1c, gO2a, gO3, gO4, and gO5 genotypes was small, we did not perform a statistical analysis of correlations between clinical features and these gO genotypes. When comparing infections involving only single genotypes, gO2b genotype had a more unfavorable outcome (p = 0.09) than the other single genotypes, but this finding was not time-related. The mixed gO genotype group compared with only single genotype group was more associated with female gender (p = 0.015), acute leukemia disease (p = 0.036), initial high level of CMV viral load (p = 0.029), and skin rash (p = 0.01) (Table 4).
Analysis of Genetic Linkage of gO/gN Variants in Non-HSCT Patients with Mixed CMV Genotype Infection
Both gO/gN genotypes were identified from 55 CMV-infected patients. Twenty-five patients with single genotypes in both gO/gN infection were found. Thirty patients were co-infected with mixed genotypes in at least one genetic locus of gO and gN. The consistent predefined linkages (described previously) of gO/gN were found as follows: gO1a/gN1 in 16.4% (9/55), gO1b/gN3a in 1.8% (1/55), gO1c/gN4c in 20% (11/55), gO2a/gN3b in 14.5% (8/55), gO3/gN4a in 20% (11/55), and gO5/gN4c in 3.6% (2/55).The linkages of gO2b/gN2 and gO4/gN4b were not found. However, novel linkages between gO/gN genotypes were found as gO3a/gN4a in 20% (11/55), gO2b/gN3b in 16.4%, gO1a/gN3b, and gO1c/gN3b in each with 12.7% (7/55), gO1a/gN4a,gO2a/gN4c, gO2b/gN1 gO2b/gN4a, andgO3a/gN4c in 10.9% (6/55), respectively. The percentage of all associations of gO/gN genotypes in CMV-infected patients is shown in Table 5.
The effects of gO/gN genotype linkages on CMV viral load are shown in Table 6. The difference between the viral load within the different linkages groups (presence and absence of linkages) was not statistically significant. However, only the presence of gO1a/gN3b linkage group showed a significantly higher viral load compared with the absence of the gO1a/gN3b linkage (p = 0.012) (Table 6).
Discussion
To our knowledge there have been limited studies on the distribution of CMV genotypes and their possible role among non-transplant patients with HM undergoing chemotherapy. And, only some studies have involved analysis of gB genotypes in this setting. Moreover, no study has addressed the distribution and the role of gN and gO genotypes in this group of patients [5].
In the present study mixed CMV infection with more than one glycoprotein genotype (gB, gN, and gO) was found in 60% of infected patients with acute leukemia and lymphoma undergoing chemotherapy. The mixed CMV gB, gN, and gO genotypes were detected in 31%, 41%, and 46.4%, respectively, of the CMV-infected patient. These results are consistent with previous studies in different population groups which underlined that co-infection with different strains is common. In fact, a high proportion of mixed CMV infections with multiple glycoprotein genotypes was noted in a variety of population groups including immunocompetent [23], patients with AIDS [22, 24], allo-HSCT recipients [25, 26], solid organ transplantation (SOT) [6, 27, 28], and infants with congenital CMV infection [29]. In contrast, a lower prevalence of mixed infections was noted in other groups of infants with congenital infection [11], AIDS [30], HSCT [12, 31], and SOT recipients [6]. In Tunisia, the distribution of gB genotypes was described in one study and it was found that the gB1and gB2 genotypes were dominant with no mixed genotypes in immunocompromised patients including transplant recipients and patients with AIDS [32]. Most of these previous studies have focused on a singular viral envelope glycoprotein gene characterized by polymerase chain reaction (PCR)-based genotyping which is relatively insensitive to multiple-strain infections [18, 33, 34]. The detection of mixed infection was also hampered by tissue-cultured viruses because of the selection of a single strain [35, 36]. However, in the current study, clinical specimens were tested directly to prevent underestimation of virus diversity and we used highly sensitive multiplex nested PCR and subsequent Sanger sequencing technology at multiple loci.
A different finding was described in previous studies with almost no mixed gN genotypes, such as in newborns with congenital infection [11] and in immunocompromised patients including SOT population [37], or patients with AIDS [30]. In striking contrast, other studies showed a higher prevalence of gN mixed infection in African children [33] and 40.2% in Chinese HSCT recipients [38]. While plenty of data are available on the gB and gN genotype, little is known about the gO genotype distribution in immunosuppressed patients.
Consistent with a previous study, mixed gO genotype was the most common genotype in allo-HSCT recipients [25]. But, this finding differs from a study by Roubalova et al. who found that the gO1 genotype was the most frequently genotype identified in allo-HSCT recipients with CMV infections [39]. The differences among studies may, in part, be due to the wide spectrum of technique sensitivity, variation in geographical and/or demographic allocation of genotypes, variability in transmission, and differences in mechanisms of pathogenesis and cause of immunosuppression [15, 27, 40].
In this study most of the patients were anti-CMV IgG seropositive before chemotherapy, and most probably viral DNAemia during chemotherapy was due to reactivation or reinfection under immunosuppressive conditions rather than primo-infection. Interestingly, analysis of different samples from the same patients at a later CMV infection episode revealed the emergence of new genotypes over time in half of patients with recurrent infections. This result may have multiple explanations. CMV reactivation or reinfection seems to occur stochastically, as has previously been shown using the murine model [41, 42]. It is possible that these patients acquired new strains during the time between the first screening and follow-up. If not, both strains may have been present at both time points but not detected in the first screening samples because of a low DNAemia of minor virus populations. Moreover, quantitative differences among the different CMV genotypes could be associated with different replication efficiencies of specific genotypes. In this context, the dominating genotype can be expected to have the most pronounced impact on the course of CMV infection. Furthermore, the relative frequencies of the individual genotypes within a patient did not remain constant over time, a finding which has also been described for Epstein–Barr virus strains in mixed infections [43]. It is also of interest to consider that genotype-specific immune responses might have an influence on the rate of replication of a particular genotype [41].
There is growing evidence that the occurrence of mixed genotype infections serves as a marker of severe or prolonged complications from CMV infections in SOT and HSCT recipients [5, 6, 25, 28, 44,45,46].Whether these observations can be extrapolated to the non-HSCT setting is yet to be elucidated. In agreement with a previous report in immunocompromised patients [5], our data showed that patients who were infected with mixed gB genotypes developed higher initial levels of CMV loads and had significantly more concomitant infection with other HHVs than patients infected with single gB genotypes. It is also interesting to note that patients infected with mixed gN genotypes were more associated with profound lymphopenia (ALC < 500/µL) and increased risk of death than patients infected with single gN genotype. In a previous study single gN1 and gN4b showed a significantly different virulence and could serve as early predictors for the progression of CMV infection in transplant patients [47].
In the present study, patients infected with gO2b genotype had more unfavorable outcome than patients infected with single gO genotypes. In comparison to single genotypes, mixed genotypes were more frequently associated with female gender, acute leukemia disease, skin rash, and the initial high level of CMV viral load. In contrast, no clear correlation existed between the gO genotypes and CMV disease in clinical isolates from different geographic regions and different disease settings [9, 16, 48, 49].
On the basis of the distinct distributions of CMV gB, gO, and gN genotypes in different clinical settings and the association between mixed genotypes and serious complications, we assumed that a single polymorphic gene as a virulence marker was not sufficient to investigate the molecular epidemiology of CMV. The virulence of CMV may depend on the synergistic action of at least two glycoproteins. Different genotypes in mixed infection can interact with each other and the interaction between co-infecting genotypes may depend on the specific genotype combination. The exact mechanism of this strain interaction for CMV has not been identified, but it has been shown in the mouse model that different murine CMV variants can interact with each other through functional trans-complementation and that this can lead to impaired virus fitness [41, 42]. The linkage between gN and gO genotypes suggests possible functional interactions between these glycoproteins which may lead to virus replication, entry, and cell tropism [11, 16, 18, 19, 47]. Interestingly, a novel linkage between gO1a and gN3b was recognized in the studied patients and was significantly associated with higher viral load unlike the other defined gO/gN linkages [18, 19]. In contrast to the previous studies [16, 22, 30], no association was observed between gO1a/gN4a, gO1a/gN1, gO3/gN4a, and viral load, which may be due to limited sample numbers in this study. Therefore, further studies are needed to ascertain the strength of this new genetic linkage and to highlight the relevance of glycoprotein genotypes as virulence factors in the progression and the severity of CMV infection in immunocompromised hosts.
This study had some limitations such as the small number of patients enrolled, the monocentric type of study, and the limited geographical area surveyed. Larger epidemiological studies and improved genotype-specific real-time PCR-based assays and high-throughput sequencing technology may reveal a high level of intrahost variability and offer strong evidence that CMV exists as a complex mixture of variants in immunocompromised patients and may also clarify more the epidemiological and the clinical impact of these variants. The genetic profile of infecting viruses and relation to clinical outcome should be investigated, taking into account the host’s innate and adaptive immune responses [4, 8, 45, 50, 51].
Conclusions
Our results provide evidence that, in non-HSCT patients with HM undergoing chemotherapy, severe clinical manifestations occur mostly in patients infected with multiple CMV genotypes. The interaction of multiple genotypes during CMV infection, more specifically by gO1a and gN3b, may play an important role in the virulence of CMV in infected non-HSCT patients.
References
Dixon SB, Lane A, O’Brien MM, et al. Viral surveillance using PCR during treatment of AML and ALL. Pediatr Blood Cancer. 2018;65:e26752.
Torres HA, Kontoyiannis DP, Aguilera EA, et al. Cytomegalovirus infection in patients with lymphoma: an important cause of morbidity and mortality. Clin Lymphoma Myeloma. 2006;6:393–8.
Phasuk N, Keatkla J, Rattanasiri S, Techasaensiri C, Anurathapan U, Apiwattanakul N. Monitoring of cytomegalovirus infection in non-transplant pediatric acute lymphoblastic leukemia patients during chemotherapy. Medicine (Baltimore). 2019;98:e14256.
Scalzo AA, Corbett AJ, Rawlinson WD, Scott GM, Degli-Esposti MA. The interplay between host and viral factors in shaping the outcome of cytomegalovirus infection. Immunol Cell Biol. 2007;85:46–54.
Coaquette A, Bourgeois A, Dirand C, Varin A, Chen W, Herbein G. Mixed cytomegalovirus glycoprotein B genotypes in immunocompromised patients. Clin Infect Dis. 2004;39:155–61.
Humar A, Kumar D, Gilbert C, Boivin G. Cytomegalovirus (CMV) glycoprotein B genotypes and response to antiviral therapy, in solid-organ-transplant recipients with CMV disease. J Infect Dis. 2003;188:581–4.
Dolan A, Cunningham C, Hector RD, et al. Genetic content of wild-type human cytomegalovirus. J Gen Virol. 2004;85:1301–12.
Rasmussen L. Molecular pathogenesis of human cytomegalovirus infection. Transpl Infect Dis. 1999;1:127–34.
Rasmussen L, Geissler A, Cowan C, Chase A, Winters M. The genes encoding the gCIII complex of human cytomegalovirus exist in highly diverse combinations in clinical isolates. J Virol. 2002;76:10841–8.
Paterson DA, Dyer AP, Milne RS, Sevilla-Reyes E, Gompels UA. A role for human cytomegalovirus glycoprotein O(gO) in cell fusion and a new hypervariable locus. Virology. 2002;293:281–94.
Pignatelli S, Lazzarotto T, Gatto MR, et al. Cytomegalovirus gN genotypes distribution among congenitally infected newborns and their relationship with symptoms at birth and sequelae. Clin Infect Dis. 2010;51:33–41.
Dieamant DC, Bonon SH, Peres RM, et al. Cytomegalovirus (CMV) genotype in allogeneic hematopoietic stem cell transplantation. BMC Infect Dis. 2013;13:310.
Arcangeletti MC, Vasile Simone R, Rodighiero I, et al. Combined genetic variants of human cytomegalovirus envelope glycoproteins as congenital infection markers. Virol J. 2015;12:202.
Chou SW, Dennison KM. Analysis of interstrain variation in cytomegalovirus glycoprotein B sequences encoding neutralization-related epitopes. J Infect Dis. 1991;163:1229–34.
Zipeto D, Hong C, Gerna G, et al. Geographic and demographic differences in the frequency of human cytomegalovirus gB genotypes 1–4 in immunocompromised patients. AIDS Res Hum Retroviruses. 1998;14:533–6.
Pignatelli S, Dal Monte P, Rossini G, et al. Human cytomegalovirus glycoprotein N (gpUL73-gN) genomic variants: identification of a novel subgroup, geographical distribution and evidence of positive selective pressure. J Gen Virol. 2003;84:647–55.
Pignatelli S, Dal Monte P, Landini MP. gpUL73 (gN) genomic variants of human cytomegalovirus isolates are clustered into four distinct genotypes. J Gen Virol. 2001;82:2777–84.
Mattick C, Dewin D, Polley S, et al. Linkage of human cytomegalovirus glycoprotein gO variant groups identified from worldwide clinical isolates with gN genotypes, implications for disease associations and evidence for N-terminal sites of positive selection. Virology. 2004;318:582–97.
Yan H, Koyano S, Inami Y, et al. Genetic linkage among human cytomegalovirus glycoprotein N (gN) and gO genes, with evidence for recombination from congenitally and post-natally infected Japanese infants. J Gen Virol. 2008;89:2275–9.
Tanaka T, Kogawa K, Sasa H, Nonoyama S, Furuya K, Sato K. Rapid and simultaneous detection of 6 types of human herpes virus (herpes simplex virus, varicella-zoster virus, Epstein-Barr virus, cytomegalovirus, human herpes virus 6A/B, and human herpes virus 7) by multiplex PCR assay. Biomed Res. 2009;30:279–85.
Tarrago D, Quereda C, Tenorio A. Different cytomegalovirus glycoprotein B genotype distribution in serum and cerebrospinal fluid specimens determined by a novel multiplex nested PCR. J Clin Microbiol. 2003;41:2872–7.
Jiang XJ, Zhang J, Xiong Y, et al. Human cytomegalovirus glycoprotein polymorphisms and increasing viral load in AIDS patients. PLoS One. 2017;12:e0176160.
Schoppel K, Kropff B, Schmidt C, Vornhagen R, Mach M. The humoral immune response against human cytomegalovirus is characterized by a delayed synthesis of glycoprotein-specific antibodies. J Infect Dis. 1997;175:533–44.
Bongarts A, Von Laer D, Vogelberg C, et al. Glycoprotein B genotype of human cytomegalovirus: distribution in HIV-infected patients. Scand J Infect Dis. 1996;28:447–9.
Vinuesa V, Bracho MA, Albert E, et al. The impact of virus population diversity on the dynamics of cytomegalovirus DNAemia in allogeneic stem cell transplant recipients. J Gen Virol. 2017;98:2530–42.
Gonzalez-Ramirez J, Uribe-Gutierrez G, Jimenez-Hernandez E, et al. Cytomegalovirus gB genotype distribution in Mexican children undergoing allogeneic bone marrow transplantation. Intervirology. 2012;55:318–20.
Khalafkhany D, Makhdoomi K, Taghizadeh Afshari A, Motazakker M. Prevalence and genotype distribution of cytomegalovirus UL55 sequence in renal transplant recipients in north west of Iran. J Med Virol. 2016;88:1622–7.
Manuel O, Asberg A, Pang X, et al. Impact of genetic polymorphisms in cytomegalovirus glycoprotein B on outcomes in solid-organ transplant recipients with cytomegalovirus disease. Clin Infect Dis. 2009;49:1160–6.
Ross SA, Novak Z, Pati S, et al. Mixed infection and strain diversity in congenital cytomegalovirus infection. J Infect Dis. 2011;204:1003–7.
Pignatelli S, Rossini G, Dal Monte P, Gatto MR, Landini MP. Human cytomegalovirus glycoprotein N genotypes in AIDS patients. AIDS. 2003;17:761–3.
Wu X, Wang Y, Xu Y, et al. Cytomegalovirus glycoprotein B genotype in hematopoietic stem cell transplant patients from China. Biol Blood Marrow Transplant. 2010;16:647–52.
Mhiri L, Meritet JF, Lebon P, Slim A. Different human cytomegalovirus glycoprotein B (gB) genotype distribution. Pathol Biol (Paris). 2008;56:39–42.
Bates M, Monze M, Bima H, et al. High human cytomegalovirus loads and diverse linked variable genotypes in both HIV-1 infected and exposed, but uninfected, children in Africa. Virology. 2008;382:28–36.
Gorzer I, Kerschner H, Jaksch P, et al. Virus load dynamics of individual CMV-genotypes in lung transplant recipients with mixed-genotype infections. J Med Virol. 2008;80:1405–14.
Sinzger C, Schmidt K, Knapp J, et al. Modification of human cytomegalovirus tropism through propagation in vitro is associated with changes in the viral genome. J Gen Virol. 1999;80(Pt 11):2867–77.
Dargan DJ, Douglas E, Cunningham C, et al. Sequential mutations associated with adaptation of human cytomegalovirus to growth in cell culture. J Gen Virol. 2010;91:1535–46.
Puchhammer-Stockl E, Gorzer I, Zoufaly A, et al. Emergence of multiple cytomegalovirus strains in blood and lung of lung transplant recipients. Transplantation. 2006;81:187–94.
Xia CS, Zhang Z. Analysis of human cytomegalovirus glycoprotein N genotypes in Chinese hematopoietic stem cell transplant recipients. Arch Virol. 2011;156:17–23.
Roubalova K, Strunecky O, Vitek A, Zufanova S, Prochazka B. Genetic variability of cytomegalovirus glycoprotein O in hematopoietic stem cell transplant recipients. Transpl Infect Dis. 2011;13:237–43.
Carraro E, Granato CF. Single human cytomegalovirus gB genotype shed in multiple sites at the time of diagnosis in renal transplant recipients. J Med Virol. 2003;70:240–3.
Gorzer I, Kerschner H, Redlberger-Fritz M, Puchhammer-Stockl E. Human cytomegalovirus (HCMV) genotype populations in immunocompetent individuals during primary HCMV infection. J Clin Virol. 2010;48:100–3.
Cicin-Sain L, Podlech J, Messerle M, Reddehase MJ, Koszinowski UH. Frequent coinfection of cells explains functional in vivo complementation between cytomegalovirus variants in the multiply infected host. J Virol. 2005;79:9492–502.
Sitki-Green D, Covington M, Raab-Traub N. Compartmentalization and transmission of multiple Epstein-Barr virus strains in asymptomatic carriers. J Virol. 2003;77:1840–7.
Barrado L, Prieto C, Hernando S, Folgueira L. Detection of glycoproteins B and H genotypes to predict the development of cytomegalovirus disease in solid organ transplant recipients. J Clin Virol. 2018;109:50–6.
Ciotti M, Cella E, Ritta M, et al. Cytomegalovirus glycoprotein B genotype distribution in Italian transplant patients. Intervirology. 2017;60:165–70.
Lisboa LF, Tong Y, Kumar D, et al. Analysis and clinical correlation of genetic variation in cytomegalovirus. Transpl Infect Dis. 2012;14:132–40.
Rossini G, Pignatelli S, Dal Monte P, et al. Monitoring for human cytomegalovirus infection in solid organ transplant recipients through antigenemia and glycoprotein N (gN) variants: evidence of correlation and potential prognostic value of gN genotypes. Microbes Infect. 2005;7:890–6.
Sowmya P, Madhavan HN. Analysis of mixed infections by multiple genotypes of human cytomegalovirus in immunocompromised patients. J Med Virol. 2009;81:861–9.
Paradowska E, Jablonska A, Studzinska M, et al. Distribution of the CMV glycoprotein gH/gL/gO and gH/gL/pUL128/pUL130/pUL131A complex variants and associated clinical manifestations in infants infected congenitally or postnatally. Sci Rep. 2019;9:16352.
Pang X, Humar A, Preiksaitis JK. Concurrent genotyping and quantitation of cytomegalovirus gB genotypes in solid-organ-transplant recipients by use of a real-time PCR assay. J Clin Microbiol. 2008;46:4004–10.
Sezgin E, An P, Winkler CA. Host genetics of cytomegalovirus pathogenesis. Front Genet. 2019;10:616.
Acknowledgements
We thank the participants of the study.
Funding
This work was funded by the Ministry of Higher Education and Scientific Research, Tunisia. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The journal’s Rapid Service Fee was funded by the authors.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work, and have given their approval for this version to be published.
Authorship Contributions
Imene Handous, Naila Hannachi, Abderrahim Khelif, and Jalel Boukadida contributed to the conception and design of research. Imene Handous, Bechir Achour, Manel Marzouk, Olfa Hazgui acquired data in the study. Imene Handous performed the experiments. Imene Handous, Naila Hannachi, Manel Marzouk, and Olfa Hazgui participated in the analysis and interpretation of data. Imene Handous and Naila Hannachi wrote the draft manuscript. Naila Hannachi, Abderrahim Khelif, and Jalel Boukadida supervised and supported the study. All authors read and approved the final manuscript.
Disclosures
Imene Handous, Naila Hannachi, Bechir Achour, Manel Marzouk, Olfa Hazgui, Abderrahim Khelif, and Jalel Boukadida have nothing to disclose.
Compliance with Ethics Guidelines
Ethical approval for the study was obtained from the Ethics Committee and Medical research of the Farhat Hached University Hospital, Sousse, Tunisia (Reference number IRB00008931). All the patients were informed about the consent of the study and signed a statement before starting the research work.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Corresponding author
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Handous, I., Hannachi, N., Achour, B. et al. Cytomegalovirus Glycoprotein Polymorphisms and Increasing Viral Load in Non-Transplant Patients with Hematological Malignancies Undergoing Chemotherapy: A Prospective Observational Study. Infect Dis Ther 10, 1549–1566 (2021). https://doi.org/10.1007/s40121-021-00457-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40121-021-00457-z