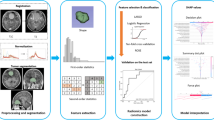
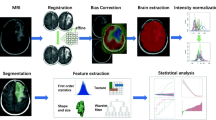
Abstract
Introduction
Conventional magnetic resonance imaging (MRI) features have difficulty distinguishing glioma true tumor recurrence (TuR) from treatment-related effects (TrE). We aimed to develop a machine-learning model based on multimodality MRI radiomics to help improve the efficiency of identifying glioma TuR.
Methods
A total of 131 patients were enrolled and randomly divided into the training set (n = 91) and the test set (n = 40). Radiomic features were extracted from the postoperative enhancement (PoE) region and edema (ED) region from four routine MRI sequences. After analyses of Spearman's rank correlation coefficient, and least absolute shrinkage and selection operator, the key radiomic features were selected to construct support vector machine (SVM) and k-nearest neighbor (KNN) models. Decision curve analysis (DCA) and receiver operating characteristic (ROC) curves were used to analyze the performance.
Results
The PoE model had a significantly higher area under curve (AUC) than the ED model (p < 0.05). Among the models constructed with a single sequence, the model using PoE regional features from CE-T1WI was superior to other models, with an AUC of 0.905 for SVM and 0.899 for KNN. In multimodality models, the PoE model outperformed the ED model with an AUC of 0.931 for SVM and 0.896 for KNN. The multimodality model, which combined routine sequences and the whole regional features, showed a slightly better performance with an AUC of 0.965 for SVM and 0.955 for KNN. Decision curve analysis showed the good clinical utility of multimodal radiomics models.
Conclusions
Multimodality radiomics can identify glioma TuR and TrE, potentially aiding clinical decision-making for individualized treatment. And edematous regions may provide useful information for recognizing recurrence.
Retrospectively Registered
2021.04.15, No:2020039.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Why carry out this study? |
Abnormal enhancement following radiotherapy is a typical secondary change that requires timely identification of true tumor recurrence from treatment-related effects and tailored treatment plan adjustment. |
Advanced imaging techniques require additional scans and impose a financial burden on patients. Using conventional MRI to construct a radiomics model may provide a universal and affordable tool. |
What was learned from the study? |
The radiomics model exhibited optimal discrimination for true tumor recurrence, with equally strong performance in the test set, and the edema area also provided rich information related to tumor recurrence. |
The radiomic model assists clinicians in efficiently and reliably evaluating patients' recurrence status and can help personalize the management and treatment of glioma patients. |
Introduction
Glioma is the most common malignant primary tumor in the brain, with a high recurrence rate and mortality rate [1]. Generally, the main treatment for newly diagnosed glioma is maximal surgical resection of the tumor lesion area [2]. Due to the infiltrative proliferation of tumor cells, these will inevitably remain in the area between the major body of the lesion and the peritumoral edema, resulting in difficulty of complete surgical resection, and the prognosis remains poor even after treatment with concurrent chemoradiotherapy [3]. After radiotherapy, the expression levels of tumor necrosis factor-α (TNF-α) are elevated in brain tissue [4], which triggers the secondary upregulation of vascular endothelial growth factor (VEGF) [5] and increases the permeability of the blood–brain barrier. Different enhancement patterns in the operative area are caused by contrast leakage during magnetic resonance imaging (MRI) enhancement [6]. This renders both true tumor recurrence (TuR) and treatment-related effects (TrE) appear occupied and abnormally enhanced, thereby making them difficult to distinguish promptly [7].
Notably, it remains challenging to differentiate TuR from TrE depending on such enhancement patterns on conventional MRI. Thus, it tends to confuse early recurrence or malignant transformation with treatment-related changes, yet their treatment strategies are distinctive. Tumor recurrence necessitates more vigorous anticancer treatment and, if possible, reoperation [8]. In contrast, TrE show the efficacy of the current treatment without the need to adjust the treatment regimen, and symptomatic treatment is sufficient. Hence, early detection of TuR is essential for adjusting clinical treatment plans [9,10,11]. Positron emission tomography–computed tomography (PET-CT) and MRI are currently the most commonly used imaging techniques for the detection of glioma recurrence. The Response Assessment in Neuro-Oncology working group and the European Association for Neuro-Oncology recommend the use of PET-CT imaging for treatment response assessment in gliomas, emphasizing that PET-CT has a higher diagnostic accuracy than MR in the identification of TuR and TrE [12]. In this study, we have focused on patients with grade II–IV gliomas, classified using the World Health Organization (WHO) grading system. These patients are often at a higher risk for postoperative recurrence, necessitating adjuvant radiotherapy or concurrent chemoradiotherapy. Close monitoring and regular follow-up imaging are vital to detect any sign of tumor growth or recurrence. In comparison with PET-CT, conventional MRI is more available in various hospitals, and does not expose patients to ionizing radiation, making it more optimal for long-term monitoring of recurrence. Our objective is to build a model based on conventional MRI sequences to identify postoperative recurrence of grade II–IV gliomas, whereas radiomics could improve diagnostic accuracy by extracting high-throughput quantitative information from the region of interest (ROI) in medical imaging [13]. Therefore, the combination of multimodality imaging and MRI radiomics is more suitable for clinical follow-up to detect early tumor recurrence, which has been previously employed in glioma recurrence studies [14,15,16]. In addition, research has demonstrated that timely and accurate distinction between early TuR and TrE may improve the life quality of individuals [17].
This study aims to develop a multimodal MRI-based machine-learning model to assist in identifying TuR from TrE, and to assess the differences between PoE and ED regional radiomics models in recognizing glioma recurrence status. We present this article in accordance with the STARD reporting checklist.
Methods
This study was approved by the institutional ethics committee of the First Affiliated Hospital of Xinxiang Medical University, China (No. 2020039). All data are anonymous, and the requirement for informed consent was therefore waived. In order to ensure the quality and reliability of the data, all pathology results and imaging findings underwent independent review. This work was performed in accordance with the 1964 Declaration of Helsinki and its later amendments. The radiomics modeling workflow are shown in Fig. 1.
Patients and Data Collection
We consecutively included 289 patients with glioma who underwent MRI and presented postoperative changes in our institution from January 2018 to April 2022. A flow chart of the study population is shown in Fig. 2. The inclusion criteria were as follows: (1) patients underwent surgical treatment; (2) pathologically confirmed grade II–IV gliomas according to the 2021 World Health Organization classification of central nervous system tumors [18]; (3) treated with radiotherapy or chemotherapy; (45) performed MRI routine and contrast-enhanced scans after adjuvant therapy; and (5) received more than 3 months of follow-up. The followings were the exclusion criteria: (1) the lesion located under the curtain or in the brain stem; (2) incomplete surgical resection; (3) large artifacts or poor-quality images did not meet diagnostic requirements; (4) had other types of central nervous system diseases; and (5) loss of follow-up. A total of 131 patients were ultimately enrolled in the primary cohort and their clinical characteristics, pathological results, and MRI data were collected. Secondary surgery or pathological puncture is the gold standard for the diagnosis of glioma recurrence, but is invasive. Therefore, we chose the Response Assessment in Neuro-Oncology (RANO) criteria to assess recurrence status based on clinical characteristics and multiple MRI findings. True tumor recurrence was identified if a patient met one of the following criteria: (1) after at least 3 months of follow-up without additional intervention, an increase of 25% or more in the area of the focal enhancing lesions or an increase in the degree of enhancement was observed on contrast-enhanced T1-weighted imaging; (2) an increase was observed in the high signal area on T2-weighted fluid attenuated inversion recovery; (2) new lesions were identified; or (4) there was progression of the mass effect or worsening clinical symptoms.
Radiomics workflow. a After acquiring medical images, preprocessing was performed, including image registration, resampling, and grayscale discretization. b Manually outline the areas of postoperative enhancement and edema separately. c Enable Laplacian of Gaussian and wavelet transform filters to extract first-order and texture features from the region of interest. d The least absolute shrinkage and selection operator was used to filter radiomics features, and then higher weighted features were further used to build machine learning models. e Repeated construction of machine-learning models based on imaging modalities and region of interest. f Assessment of the model's classification performance using receiver operating characteristic curve and decision curves
The primary cohort, which consisted of 72 patients with TuR and 59 patients with TrE, was then randomized in a 7:3 ratio to the training set (n = 90) and test set (n = 41). All patients were scanned on 3.0 T scanners and the following four routine sequences were included in the study: axial T1-weighted image (T1WI), T2-weighted image (T2WI), T2-weighted fluid attenuated inversion recovery (T2-FLAIR), and contrast-enhanced T1-weighted image (CE-T1WI). Detailed information on the MR machine and sequence parameter is provided in Table S1 in the Electronic Supplementary Material (ESM).
Image Processing and ROI Delineation
The study included MRI sequences that were acquired no more than 3 months after the patient had received therapy. In order to decrease discrepancies in the grayscale values of an image caused by different scanning equipment, the signal intensities were processed with Min–Max normalization. Then, the voxels were resampled to 1 × 1 × 1 mm3 by using the nearest neighbor interpolation to discretize the grayscale data by 25 bin widths. We utilized rigid and affine transformations (ANTS, v.2.4.3, RRID:SCR_004757) to align CE-T1WI, T2-FLAIR, and T2WI with T1WI before delineating the ROI [19].
Under the guidance of a chief radiologist with 30 years of experience in neurological diagnosis, two radiology graduate students manually outlined ROIs layer by layer using the segmentation tool in 3D Slicer (v.4.13.0, RRID:SCR_005619), primarily based on CE-T1WI and T2-FLAIR to outline postoperative enhancement (PoE) and edema region (ED), while multiple sequences were cross-referenced to define the boundaries. In a boundary controversy, the chief physician was consulted for a consensus. Two readers re-outlined the ROI 1 month later and kept good agreement (ICC > 0.75). The patient's disease status was not known to either reader during the outlining of the ROI.
Extraction of Radiomics Features
Radiomics features were extracted automatically by the open-source software Pyradiomics (v.3.0.1, https://pyradiomics.readthedocs.io/en/latest) [20]. Three types of features were extracted from each sequence, including shape features (n = 14), first-order intensity features (n = 18), and texture features (n = 75). Firstly, 14 shape features were extracted from each original image, and then 13 transformed images were obtained after Wavelet and Laplacian of Gaussian (LoG; sigma: 1, 2, 3, 4, 5) filters. First-order features and texture features were extracted from all images. Ultimately, a total of 1316 [14 + (13 + 1) × (18 + 75)] features were derived from each sequence. The ICC was utilized to measure the consistency of the features extracted by the two readers. Features with ICC > 0.75 were retained to ensure the study stability. All collected features adhere to the Imaging Biomarker Standardization Initiative [21].
Data Processing and Analysis
Initially, all missing data were removed. Then, the data of all radiomics features were preprocessed with Z-score normalization and utilized Spearman's rank correlation coefficient to remove redundant features with strong correlations. For correlation coefficients greater than 0.9, only one of the features was kept, and the other was dismissed. The remaining features were then further filtered by using the ten-fold cross-validated least absolute shrinkage and selection operator (LASSO) regression to select features corresponding to the λ values that had the best prediction and the least model bias.
After ranking all features with non-zero coefficients, key features with high weights were selected. Features were categorized into 15 groups based on the imaging modality and ROI type. Each group's key features were then used to develop support vector machine (SVM) and k-nearest neighbor (KNN) models.
Model Development and Validation
In the training set, SVM and KNN classifiers were developed with radiomics features extracted from the PoE and ED regions. Five-fold cross-validation was used on all models to ensure classification performance. The following three strategies will assist in identifying and validating models with good classification efficacy for TuR.
-
I.
Regional-level (PoE vs. ED vs. Whole). To evaluate whether the subset of features inside different regions could provide valid information, the performance in recognizing TuR was separately assessed by using features from PoE and ED. We further constructed a multimodality model with the features from both areas to determine whether the multi-area features could improve the identification performance.
-
II.
Sequence-level (T1WI vs. CE-T1WI vs. T2-FLAIR vs. T2WI vs. Multimodality). Every sequence was analyzed with features from the same region to determine which one has the best performance in identification. Then, multimodality sequences were combined and compared with a single sequence to take advantage of MRI multiparametric imaging.
-
III.
Model-level (SVM vs. KNN). All models were based on SVM or KNN classifiers and were compared. Consequently, the optimal feature combination and classifier model was developed and further validated in the test set.
In the test set, receiver operating characteristic (ROC) curves were used to evaluate the accuracy, sensitivity, specificity and AUC of the model. The clinical applicability of the model was then accessed through decision curve analysis (DCA) [22], which calculated the net benefit of the model at different thresholds.
Statistical Analysis
SPSS 19.0 software (SPSS, Chicago, IL, USA) was used for statistical analysis. Continuous variables in the clinical characteristic were analyzed by using Student's t test or Mann–Whitney U test, while categorical variables were analyzed by using the chi-square (χ2) test. Univariate logistic regression was used to assess the relationship between recurrence status and the clinical characteristics of patients. ROC analysis was used to evaluate the diagnostic effectiveness of the SVM and KNN classifiers. DCA was also utilized to determine whether the prediction model has clinical application value on the Python 3.7.12 platform. Analyses were deemed statistically significant if p < 0.05, and all tests were two-tailed.
Results
Characteristics of Patients
The clinical characteristics of all patients in the training and test sets are presented in Table 1. According to the RANO criteria [23], 72 patients were considered to have TuR, and 59 patients were considered to have TrE. In terms of clinical characteristics, there were no significant differences between the training and test sets (p > 0.05).
Feature Selection and Modeling
Firstly, the feature results extracted by the two readers were evaluated for consistency by using ICC. Features with ICC value greater than 0.75 were retained. There were 3441 features in the PoE area and 3424 in the ED area. After Spearman correlation analysis, 794 features from the PoE and 736 features from the ED region were suggested to have no redundant information. Subsequently, we used the LASSO algorithm to carry out dimensionality reduction and to select high-weighted features for the classification model. However, considering that the number of features used for the final model should not exceed one-tenth of the sample size [24], 18 (10/8, represents that ten features were from the PoE region and eight features from the ED region) features of T1WI, 17 (9/8) features of CE-T1WI, 17 (9/8) features of T2WI, and 20 (10/10) features of FLAIR were chosen to construct the SVM and KNN models. Finally, we obtained a dataset with 72 key radiomics features that consist of 38 features from the PoE area and 34 from the ED area. These key features are shown in Table S2 in the ESM.
Performance of Models
First, a region-level comparison was performed among radiomics features from the PoE region, the ED region, and the whole region. In the training set, models constructed with features from PoE or ED region alone showed good classification performance (p < 0.01). For the model with features from the whole region, the AUC of T2WI was the most improved, from 0.848 ± 0.075 (mean ± 95% CI) to 0.942 ± 0.041. The AUCs of other models also increased with various degrees (Table S3 in the ESM). In the test set, the highest AUC was 0.905 ± 0.115 for the PoE of CE-T1WI and 0.842 ± 0.119 for the ED of T2WI in the single sequence. For the model with the whole region feature, it had the best diagnostic performance in both SVM and KNN, with AUCs of 0.939 ± 0.111 and 0.923 ± 0.111 in CE-T1WI (Fig. 3), respectively, which was consistent with the performance of the training set. From each sequence, the feature with the highest weight was selected for visualization. Figure 4 illustrates the feature maps of the four features used in the machine-learning model to describe local brain tissue differences.
ROC analysis of the classification performance of each single sequence (a–d) and multimodal radiomics (e, f) models based on SVM and KNN classifiers. SVM support vector machine, KNN k-nearest neighbor, PoE features from the postoperative enhancement regions, ED features from the edematous regions, Whole includes features of PoE and edematous regions, Multimodality combine all sequences
Representative MRI and radiomics feature maps. Conventional MRI images (a) and radiomic feature maps (c) of patients with true tumor recurrence. Conventional MRI images (b) and radiomic feature maps (d) of patients with treatment-related effects. Radiomics feature maps from left to right are T1WI wavelet-HLL ngtdm Contrast, CE-T1WI log-sigma-5–0-mm-3D firstorder Range, T2WI log-sigma-1–0-mm-3D firstorder Kurtosis and T2-FLAIR wavelet-LLH glrlm GrayLevelVariance. The area within the yellow line represents the region of postoperative enhancement, while the area between the red and yellow lines represents the region of edema
The second was a sequence-level comparison by using the same regional features of different sequences to construct models. In the training set, the model with multi-modality features reached the highest AUC (0.994 ± 0.010). In the test set, the highest AUC was 0.965 ± 0.069 and 0.955 ± 0.068 for the SVM- and KNN-based multimodality models, respectively. The highest AUCs for T1WI, CE-T1WI, T2-FLAIR, and T2WI were 0.851 ± 0.145, 0.939 ± 0.111, 0.883 ± 0.101, and 0.917 ± 0.087, respectively. Among the models constructed with single sequence, CE-T1WI showed the best performance.
Finally, a model-level comparison was conducted to determine which of SVM and KNN models has better diagnostic efficacy. The classification efficacy of SVM models in the training set were all higher than those of KNN models. In the test set, the T2-FLAIR model in the PoE region and the T1WI, CE-T1WI, and T2WI models in the ED region based on KNN outperformed the SVM model, while all other SVM models outperformed the KNN models (Table 2). Decision curves of three multimodal models based on SVM were compared. The results indicated that, if the threshold probability of clinical decision was greater than 0.05, all models added more benefit than the scheme of treat-none or treat-all patients. When the threshold probability was over 0.31 for identifying recurrence, the whole region model brought more benefit than either the PoE or ED model alone in most cases (Fig. 5).
Discussion
In this study, we developed and validated a machine learning model based on radiomics features for detecting postoperative tumor recurrence to aid in the timely therapeutic adjustment of treatment strategies, which may benefit some patients. Notably, results showed that the PoE region provides abundant information for identifying early tumor recurrence, and that CE-T1WI contains more biological information to accurately reflect regional tissue heterogeneity. Therefore, the multimodality based on the whole region best distinguished TuR from TrE, while the CE-T1WI model based on the PoE region only ranked second. Although the model based on the ED region showed a slightly inferior performance in classification (p < 0.05), it still achieved 82.5% accuracy, suggesting that the edema region could also provide reliable information.
Moreover, most of the key features selected in the tumor-enhancing and edema regions are non-duplicative, indicating that the information provided by these two types of features is relatively independent. Additionally, most of the features were derived from CE-T1WI and T2WI. Owing to the utilization of exogenous contrast agents with CE-T1WI and artificial contrast increase, the information richness was amplified and allow us to assess the extent of blood–brain barrier impairment. T2WI depicts the cellular proliferation state of neighboring tissues via free water reaction. However, T1WI and T2-FLAIR were underrepresented in terms of feature count, due to T1WI's scarcity of heterogeneous data as a structural image and T2-FLAIR's liquid inhibition resulting in partial information loss. Our findings corroborate this observation. After visualization of the imaging data, we noted that the visual patterns seen by the naked eye were consistent with the corresponding MRI findings, thereby improving our comprehension of the physiological and pathological connotations of these features. These visualizations can serve as an invaluable guide for research on specific feature classes or types, which can enhance their interpretability and replicability.
Our study indicated that both the KNN model and the SVM model could identify tumor recurrence based on the features of edema. This result confirmed our hypothesis that the texture information of the edema area could potentially provide valid information for detecting TuR. Furthermore, the SVM classifier demonstrated exceptional robustness in our study, with satisfactory performance in the majority of studies. This finding suggests that the SVM model has a broad range of applications and can facilitate the development of dependable models. Specifically, our observation implies that the SVM model has a wide variety of applications and can aid in creating stable models.
Additionally, it may assist patients with postoperative glioma with risk stratification and survival prediction [25, 26]. Recent studies [27, 28] have shown that the recurrence risk of a single tumor was 56% within 1.0 cm of the initial lesion margin and 28% within 2.0 cm. Since peritumoral edema is a typical symptom of glioma and the region is prone to recurrence [29], we intentionally included the region of edema in contrast to previous studies [30, 31]. Furthermore, by merging the multimodality imaging features of MRI, the best features from each sequence were chosen independently to ensure a comprehensive modeling. Global textural features were also used, which may reflect the heterogeneity and aggressiveness of the tumor. Currently, research on voxel-based radiomics features has been conducted [32,33,34], providing the ability to predict the risk of white matter recurrence and the site of recurrence for glioma patients. This may further advance the application of radiomics models in clinical settings.
In both the training and test sets, our model is able to adequately distinguish between TuR and TrE, and has diagnostic robustness. This allows clinicians to identify patients who are most likely to benefit from additional surgery. Better diagnostic accuracy from multimodal models could help doctors pinpoint which patients will benefit from customized therapy for glioma recurrence. In order to quantify the clinical utility, we calculated the models' net benefit at different threshold probabilities through DCA. The results confirmed that all three SVM-based multimodal models provide varying degrees of clinical usefulness at threshold probabilities of 0.05–0.89, and the whole-region features have better clinical utility in most cases. This may help clinical doctors develop personalized treatment plans, so that some patients can benefit from extra treatment.
As IDH is a biomarker highly correlated with glioma prognosis, we attempted to incorporate patients' IDH mutation status in this study. However, some patients did not receive adequate immunohistochemistry, resulting in it not showing statistically significant differences in the primary cohort. Therefore, we constructed the final model based on radiomics features only, which has shown a high accuracy. To enhance the model's generalizability, the ideal course of action would be to develop a standardized scanning protocol based on a larger dataset. This approach will help ensure that the model is applicable to a greater range of clinical scenarios.
With the development of MRI techniques, advanced MRI sequences can provide more structural and metabolic information of the lesion, and have yielded satisfactory results in studies to identify suspected recurrent gliomas [35,36,37]. The combination of advanced MRI and radiomics could be used to further explore the potential information within the images. However, few studies have been conducted to identify glioma recurrence.
Despite some success in tumor diagnosis and evaluation, radiomics methods face several challenges that hinder their clinical application. These challenges include establishing large-scale imaging databases, implementing standardized image analysis algorithms, and ensuring the security and confidentiality of data. For ensuring the reproducibility and clinical utility of the results, independent validation of the model is essential. This study has the certain limitations. First, since the investigation is retrospective, there may be selection bias. Second, the majority of patients with postoperative glioma recurrence prefer radiotherapy or chemotherapy, and only a small number of patients receive secondary surgery or pathological puncture confirmation, which may lead to inconsistent results as judged by RANO. Third, we used single-center data, so the sample size needs to be expanded. Hence, multicenter research and larger samples are required.
Conclusions
Multimodality radiomics confirmed that features of the enhanced and edematous regions in the operative area can be used to identify early glioma recurrence. This may facilitate timely clinical intervention and treatment to benefit some patients.
References
Ostrom QT, Cioffi G, Gittleman H, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2012–2016. Neuro Oncol. 2019;21:v1–100.
Xu S, Tang L, Li X, Fan F, Liu Z. Immunotherapy for glioma: current management and future application. Cancer Lett. 2020;476:1–12.
Wallner KE, Galicich JH, Krol G, Arbit E, Malkin MG. Patterns of failure following treatment for glioblastoma multiforme and anaplastic astrocytoma. Int J Radiat Oncol Biol Phys. 1989;16:1405–9.
Wilson CM, Gaber MW, Sabek OM, Zawaski JA, Merchant TE. Radiation-induced astrogliosis and blood-brain barrier damage can be abrogated using anti-TNF treatment. Int J Radiat Oncol Biol Phys. 2009;74:934–41.
Jain RK. Tumor angiogenesis and accessibility: role of vascular endothelial growth factor. Semin Oncol. 2002;29:3–9.
Sharma HS, Muresanu DF, Castellani RJ, et al. Pathophysiology of blood-brain barrier in brain tumor. Novel therapeutic advances using nanomedicine. Int Rev Neurobiol. 2020;151:1–66.
Liu J, Li C, Chen Y, et al. Diagnostic performance of multiparametric MRI in the evaluation of treatment response in glioma patients at 3T. J Magn Reson Imaging. 2020;51:1154–61.
Salvati M, Pesce A, Palmieri M, Floriana Brunetto GM, Santoro A, Frati A. The role and real effect of an iterative surgical approach for the management of recurrent high-grade glioma: an observational analytic cohort study. World Neurosurg. 2019.
Soni N, Ora M, Mohindra N, Menda Y, Bathla G. Diagnostic performance of PET and perfusion-weighted imaging in differentiating tumor recurrence or progression from radiation necrosis in posttreatment gliomas: a review of literature. Am J Neuroradiol. 2020;41:1550–7.
Kickingereder P, Burth S, Wick A, et al. Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology. 2016;280:880–9.
Easaw JC, Mason WP, Perry J, et al. Canadian recommendations for the treatment of recurrent or progressive glioblastoma multiforme. Curr Oncol. 2011;18:e126–36.
Albert NL, Weller M, Suchorska B, et al. Response Assessment in Neuro-Oncology working group and European Association for Neuro-Oncology recommendations for the clinical use of PET imaging in gliomas. Neuro Oncol. 2016;18:1199–208.
Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–6.
Shim KY, Chung SW, Jeong JH, Hwang I, Park CK, Kim TM, Park SH, Won JK, Lee JH, Lee ST, Yoo RE, Kang KM, Yun TJ, Kim JH, Sohn CH, Choi KS, Choi SH. Radiomics-based neural network predicts recurrence patterns in glioblastoma using dynamic susceptibility contrast-enhanced MRI. Sci Rep. 2021;11:9974.
Chougule T, Gupta RK, Saini J, et al. Radiomics signature for temporal evolution and recurrence patterns of glioblastoma using multimodal magnetic resonance imaging. NMR Biomed. 2022;35: e4647.
Lohmann P, Elahmadawy MA, Gutsche R, et al. FET PET radiomics for differentiating pseudoprogression from early tumor progression in glioma patients post-chemoradiation. Cancers (Basel). 2020;12.
Hu LS, Hawkins-Daarud A, Wang L, Li J, Swanson KR. Imaging of intratumoral heterogeneity in high-grade glioma. Cancer Lett. 2020;477:97–106.
Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23(8):1231–51.
Avants BB, Tustison N, Gang SJOI. Advanced normalization tools (ANTS). 2008;1–35.
van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–7.
Zwanenburg A, Vallières M, Abdalah MA, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295:328–38.
Vickers AJ, Cronin AM, Elkin EB, Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak. 2008;8:53.
Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28:1963–72.
Wang W, Peng Y, Feng X, et al. Development and validation of a computed tomography-based radiomics signature to predict response to neoadjuvant chemotherapy for locally advanced gastric cancer. JAMA Netw Open. 2021;4: e2121143.
Prasanna P, Patel J, Partovi S, Madabhushi A, Tiwari P. Radiomic features from the peritumoral brain parenchyma on treatment-naïve multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: preliminary findings. Eur Radiol. 2017;27:4188–97.
Kim JY, Yoon MJ, Park JE, Choi EJ, Lee J, Kim HS. Radiomics in peritumoral non-enhancing regions: fractional anisotropy and cerebral blood volume improve prediction of local progression and overall survival in patients with glioblastoma. Neuroradiology. 2019;61:1261–72.
Lo Greco MC, Milazzotto R, Liardo RLE, et al. Relapsing high-grade glioma from peritumoral zone: critical review of radiotherapy treatment options. Brain Sci. 2022;12.
Tu Z, Xiong H, Qiu Y, Li G, Wang L, Peng S. Limited recurrence distance of glioblastoma under modern radiotherapy era. BMC Cancer. 2021;21:720.
Seidel C, Dörner N, Osswald M, et al. Does age matter? A MRI study on peritumoral edema in newly diagnosed primary glioblastoma. BMC Cancer. 2011;11:127.
Gao XY, Wang YD, Wu SM, et al. Differentiation of treatment-related effects from glioma recurrence using machine learning classifiers based upon pre-and post-contrast T1WI and T2 FLAIR subtraction features: a two-center study. Cancer Manag Res. 2020;12:3191–201.
Wang ZH, Xiao XL, Zhang ZT, He K, Hu F. A radiomics model for predicting early recurrence in grade II gliomas based on preoperative multiparametric magnetic resonance imaging. Front Oncol. 2021;11: 684996.
Yang Y, Han Y, Zhao S, et al. Spatial heterogeneity of edema region uncovers survival-relevant habitat of glioblastoma. Eur J Radiol. 2022;154: 110423.
Dasgupta A, Geraghty B, Maralani PJ, et al. Quantitative mapping of individual voxels in the peritumoral region of IDH-wildtype glioblastoma to distinguish between tumor infiltration and edema. J Neurooncol. 2021;153(2):251–61.
Chougule T, Gupta RK, Saini J, et al. Radiomics signature for temporal evolution and recurrence patterns of glioblastoma using multimodal magnetic resonance imaging. NMR Biomed. 2022;35(3): e4647.
Hou H, Diao Y, Yu J, et al. Differentiation of true progression from treatment response in high-grade glioma treated with chemoradiation: a comparison study of 3D-APTW, 3D-PcASL imaging and DWI. NMR Biomed. 2022;e4821.
Jiang S, Guo P, Heo HY, et al. Radiomics analysis of amide proton transfer-weighted and structural MR images for treatment response assessment in malignant gliomas. NMR Biomed. 2022;e4824.
Feng A, Yuan P, Huang T, Li L, Lyu J. Distinguishing tumor recurrence from radiation necrosis in treated glioblastoma using multiparametric MRI. Acad Radiol. 2022;29:1320–31.
Acknowledgements
The authors acknowledge Liu-Yue Zhao for writing assistance. Liu-Yue Zhao did not receive funding for this support. We also extend our gratitude to all the study participants and all the members of the MRI and Neurology departments in our hospital for their invaluable assistance in this study.
Funding
This study was supported by the Henan Medical Science and Technology Research Program (Grant number LHGJ20210498) and the Research Innovation Program of the First Affiliated Hospital of Xinxiang Medical University [Grant numbers YJSCX202295Y], the rapid service see was funded by the authors.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Authors’ contributions
Study concept and design: Jinfa Ren, Xiaoyang Zhai and Dongming Han; analysis and interpretation of data: Jinfa Ren, Xiaoyang Zhai, Huijia Yin, Fengmei Zhou, Ying Hu, Kaiyu Wang, Ruifang Yan and Dongming Han; drafting of the manuscript: Jinfa Ren, Xiaoyang Zhai, Huijia Yin and Kaiyu Wang; data collection: Jinfa Ren, Xiaoyang Zhai, Ruifang Yan and Dongming Han; critical revision of the manuscript for important intellectual content: Jinfa Ren, Xiaoyang Zhai, Huijia Yin, Fengmei Zhou, Ying Hu and Dongming Han.
Disclosures
Jinfa Ren, Xiaoyang Zhai, Huijia Yin, Fengmei Zhou, Ying Hu, Kaiyu Wang, Ruifang Yan and Dongming Han declare that they have no competing interests.
Compliance with ethics guidelines
This study was approved by the institutional ethics committee of the First Affiliated Hospital of Xinxiang Medical University (No.: 2020039). All data are anonymous, and the requirement for informed consent was therefore waived. This work was performed in accordance with the 1964 Declaration of Helsinki and its later amendments.
Data availability
All data generated or analyzed during this study are available from the corresponding author on reasonable request.
Open access
This article is licensed under a Creative Commons Attribution- NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
Author information
Authors and Affiliations
Corresponding author
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Ren, J., Zhai, X., Yin, H. et al. Multimodality MRI Radiomics Based on Machine Learning for Identifying True Tumor Recurrence and Treatment-Related Effects in Patients with Postoperative Glioma. Neurol Ther 12, 1729–1743 (2023). https://doi.org/10.1007/s40120-023-00524-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40120-023-00524-2