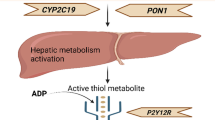
Abstract
Introduction
Trinidad & Tobago has the highest prevalence of cardiovascular disease (CVD) in the Caribbean and clopidogrel is a ubiquitously used treatment. Yet, the extent of genetically mediated clopidogrel resistance is unknown. To determine this, we investigated whether the association between CYP2C19*2 and CYP2C19*3 genetic variants and clopidogrel resistance holds, and calculated the frequencies of these in the Trinidadian CVD population.
Methods
Demographic data, clinical data, and a saliva sample were collected under informed consent from 22 patients with CVD on dual anti-platelet therapy whose biochemical resistance to clopidogrel is known, and a further 162 patients accessing the main public CVD clinic in Trinidad and who are either currently being treated or are likely to be treated with clopidogrel. A polymerase chain reaction (PCR) and restriction enzyme digestion procedure was used to genotype each patient for the CYP2C19*2 and CYP2C19*3 allelic variants. Genotype was compared to known clopidogrel resistance in the 22 patients, and to disease status and clopidogrel usage in the larger cohort.
Results
CYP2C19*2 genotype was concordant with clopidogrel resistance. CYP2C19*2 was detected in 61.1% (99/162) of patients and CYP2C19*3 was undetected. Clopidogrel was the most prescribed antiplatelet therapy (42%). A total of 120 people presented with coronary artery disease (CAD) and 52.5% of these (n = 63/120) are currently prescribed clopidogrel. 63.5% (40/63) of patients with CAD who are prescribed clopidogrel carry the CYP2C19*2 allele; ten homozygous and 30 heterozygous. Indian patients comprised 65% of the cohort and were four times more likely to carry the CYP2C19*2 allele than African patients.
Conclusions
A large proportion of Trinidadian patients with CVD who are prescribed or may be prescribed clopidogrel carry genetic variants associated with clopidogrel resistance. These results emphasize the clinical need for further investigation into whether CYP2C19*2 genotype should guide clopidogrel use for the cardiovascular disease population in Trinidad & Tobago.
A slide deck is available for this article.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Why carry out this study? |
The Caribbean is undergoing an epidemiological transition and faces the highest burden from non-communicable diseases among developing countries in the Americas. |
Trinidad and Tobago has the highest prevalence of cardiovascular disease (CVD) in the Caribbean and clopidogrel is a ubiquitously used treatment. |
The extent of genetically mediated clopidogrel resistance in the CVD population is unknown. |
What was learned from the study? |
There is a high proportion of patients with CVD in Trinidad and Tobago who are prescribed clopidogrel and carry genetic variants associated with clopidogrel resistance. |
CYP2C19 genotype-guided antiplatelet therapy may be applicable and beneficial to this population, and warrants further investigation. |
Digital Features
This article is published with digital features, including a slide deck, to facilitate understanding of the article. To view digital features for this article, go to https://doi.org/10.6084/m9.figshare.24851268.
Introduction
It is now well established that genetic variants in genes involved in drug metabolism are key modulators of variable drug response among individuals. Pharmacogenomics involves the use of this genetic data to inform the selection of the most optimally effective drug and dosage for a patient. Ultimately, pharmacogenomics promises a safer, faster, and more cost-effective treatment regime than the status quo. However, the occurrence of these genetic variants and their effects on drug metabolism have only been explored and established in a select few human populations thus far. In order to reduce health disparities, ensure that all populations benefit from genomic advances, and achieve equitable implementation of precision medicine and its associated efficiencies, it is imperative to investigate these phenomena in diverse and traditionally understudied populations [15, 27].
Caribbean populations are largely comprised of the descendants of those brought to the region via the trans-Atlantic slave trade in the 17th and 18th centuries and subsequent colonial indentureship schemes of the 19th century. Centuries and several generations later, these descendants are experiencing a non-communicable disease (NCD) epidemiological transition with the Caribbean region facing the highest burden from NCDs among developing countries in the Americas. NCDs are the leading cause of death and account for more than 70% of mortality in this region, on par with the global average [22]. Trinidad and Tobago (T&T) is an ethnically diverse country of 1.3 million people comprised of ~ 1/3 Indian (South Asian), ~ 1/3 Black (Afro-Caribbean/African), and ~ 1/3 mixed along with small populations of Chinese, Syrian/Lebanese, and Whites. NCDs account for 81% of all deaths in T&T with cardiovascular disease (CVD) accounting for 33% of all mortality (WHO, 2018). There is observed ethnic variability in those who suffer from CVD with the highest rates of CVD occurring in the Indian subpopulation [3]. Yet, under the universal health system, which includes free access to most drugs for the population, most patients with CVD are treated the same regardless of ancestry, and genetic background is not formally considered in the treatment algorithm. We set out to investigate whether pharmacogenomics may play a role in increasing the efficiency of CVD treatment using the common drug clopidogrel in this country.
Clopidogrel is prescribed as an oral inhibitor of platelet aggregation to prevent the formation of dangerous blood clots and subsequent morbid events in patients diagnosed with cardiovascular disease [9]. It is one of the most commonly prescribed medications because it can improve clinical outcomes for a broad range of cardiovascular conditions [10, 31]. The enzyme most implicated in the metabolic activation of clopidogrel is CYP2C19 [21]. There are several allelic variants of CYP2C19, categorized as either loss-of-function or gain-of-function [30]. CYP2C19*2 and CYP2C19*3 are the most studied loss-of-function variants, which both produce truncated inactive enzymes [5, 17, 19]. Specific CYP2C19 genotypes are associated with poor (CYP2C19*2/*2, CYP2C19*2/*3 or CYP2C19*3/*3), intermediate (CYP2C19*1/*2, CYP2C19*2/*17, CYP2C19*1/*3 or CYP2C19*3/*17), rapid (CYP2C19*1/*17), and ultra-rapid (CYP2C19*17/*17) CYP2C19 metabolism compared to the normal CYP2C19*1/*1 genotype associated with normal CYP2C19 metabolism [12].
Clopidogrel therapy is highly problematic when prescribed to patients carrying loss-of-function CYP2C19 variants. It is associated with a heightened risk of major adverse cardiovascular events (MACE) as a direct result of impaired CYP2C19 metabolic activation of clopidogrel [6, 9, 16]. Such patients are susceptible to a diminished quality of life and their physical, emotional, and financial well-being are impacted (Boden 2012). Importantly, where dual antiplatelet therapy is required and the patient has a genotype associated with resistance to clopidogrel, there are alternative drugs such as prasugrel or ticagrelor which can be used, if there is no contraindication [12].
The prevalence of intermediate and poor CYP2C19 metabolism varies considerably across ethnic groups and geographic regions [7, 33]. However, CYP2C19 genetic polymorphism prevalence data are lacking in Caribbean populations and their diaspora [2]. A review article of 52,181 healthy participants from 138 research articles estimated that the prevalence of intermediate and poor CYP2C19 metabolism is higher in Asian populations at 57.7%, than in African, European, and Middle Eastern populations at 32.2%, 24.8% and 30.6%, respectively [7, 9], with the CYP2C19*2 variant primarily implicated. This trend was also observed in the Trinidadian population in a pilot genetic study [18] and a pilot clinical study [25]. In the genetic study, 100 non-coronary heart disease patients were genotyped for the CYP2C19*2 variant. The prevalence of genotypes associated with intermediate and poor CYP2C19 metabolism was found to be higher in the South Asian population at 47.5% 95% confidence interval (CI)(CYP2C19*1/*2: 20%; CYP2C19*2/*2: 27.5%), than in the Caribbean Black and mixed ethnicity populations at 22.5% 95% CI (CYP2C19*1/*2: 10%; CYP2C19*2/*2: 12.5%) and 45% 95% CI (CYP2C19*1/*2: 30%; CYP2C19*2/*2: 15%), respectively [18]. In the clinical study, the clopidogrel metabolizer status of 40 patients with angina was determined using an on-treatment platelet reactivity assay. A significant South Asian predilection of 5.4 95% CI (1.18–24.66), p value 0.029 to decreased clopidogrel response in patients with cardiovascular disease was found [25]. Collectively, these data suggest that CYP2C19 genotype may be an important clinical indicator of clopidogrel efficacy in Trinidad, the island with the highest reported prevalence of cardiovascular disease in the Caribbean [22].
In this study, we investigated the relationship between biochemically determined phenotypic clopidogrel resistance and CYP2C19 loss-of-function genetic variants in a cohort of 22 patients with cardiovascular disease on dual anti-platelet therapy from the previously described clinical study [25]. We then investigated the allele and genotype frequencies of CYP2C19*2 and CYP2C19*3 variants in 162 patients with cardiovascular disease in Trinidad who are either currently being treated or are likely to be treated with clopidogrel. We discuss the implications of our findings on the treatment practices for patients with cardiovascular disease in the public health sector in T&T.
Methods
Ethical Approval
This study was performed in accordance with the Helsinki Declaration of 1964 and its later amendments. Ethical approval was obtained from the Campus Research Ethics Committee of The University of the West Indies, St. Augustine, Trinidad and Tobago (Reference Number CEC610/05/18). All participants were recruited under informed consent.
Study Population
The study population comprised adult patients registered at the Cardiac Catheterization Laboratory (CCL) at the Eric Williams Medical Sciences Complex (EWMSC), T&T at the time of sample collection. The CCL is affiliated with The University of the West Indies and receives patients with cardiovascular disease (CVD) in T&T with an estimated catchment population of over 1 million persons. A pilot cohort of 22 participants was recruited based on re-contact after participation in a previous study [25] comprising 40 participants. The remaining 18 participants were either deceased (n = 7) or unreachable (n = 11). The extent of platelet aggregation in the presence of the P2Y12 inhibitor drug clopidogrel (Plavix) was previously measured in these participants using the VerifyNow assay where high on-treatment platelet reactivity was defined as P2Y12 reaction units (PRU) > 208. As such, their biochemical clopidogrel-resistance phenotype is known. However, they were not previously genotyped. The main cohort of an additional 162 participants was newly recruited.
Sample Collection and DNA Extraction
From April 2018 to June 2018, a saliva sample was collected from each of 22 participants in the pilot cohort using the Oragene Discover OGR-500 saliva collection kit according to the manufacturer’s protocol (DNA Genotek Inc., Ottawa, Canada). From July 2019 to August 2019, data relating to age, sex, ethnicity, medical history, and prescribed medications were collected from each newly recruited participant by means of a closed-ended questionnaire. A saliva sample was also collected from each participant as above. Data and samples were de-identified for subsequent analysis. Genomic DNA was extracted from saliva samples using the prepIT L2P Manual Purification kit (DNA Genotek Inc., Ottawa, Canada) according to the manufacturer’s protocol. DNA quantity and quality were determined by standard methods.
Genotyping Assay for the CYP2C19*2 and CYP2C19*3 Variants
Each participant was genotyped for the CYP2C19*2 and CYP2C19*3 variants using a previously published two-step procedure [5, 17]. First, the CYP2C19*2 and CYP2C19*3 genetic loci were amplified by polymerase chain reaction (PCR) using the following primers; CYP2C19*2: 5'-AATTACAACCAGAGCTTGGC-3' and 5'-TATCACTTTCCATAAAAGCAAG-3' [17] and CYP2C19*3: 5'-AAATTGTTTCCAATCATTTAGCT-3' and 5'-ACTTCAGGGCTTGGTCAATA-3' [5]. Reactions of 25 μl were performed containing 12.5 μl 2X Taq PCR premix (Bioland Scientific LLC., Paramount, CA, USA), 0.5 μl forward and 0.5 μl reverse primer (Integrated DNA Technologies, Coralville, IA, USA), 200-ng DNA template, and nuclease-free water. Target sequences were amplified as follows: initial denaturation at 94 °C for 5 min; 35 cycles of denaturation at 94 °C for 45 s, annealing at 53 °C for 40 s, and extension at 72 °C for 30 s; then final extension at 72 °C for 5 min [32]. Then, PCR amplicons were digested in a 10.5-μl reaction mixture containing 10 μl PCR product and 0.5 μl restriction enzyme (Promega, Madison, WI, USA) for 60 min. For the CYP2C19*2 variant, SmaI was used at 25 °C, and for the CYP2C19*3 variant, BamHI was used at 37 °C. All reactions were performed using the Biometra TOne Thermocycler (Analytik, Jena, Germany). The products were then refrigerated at 4 °C for 30 min to stop the reaction before they were electrophoresed in a 3% agarose gel at 80 V for 70 min, along with a 100-bp DNA ladder (Bioland Scientific LLC., Paramount, CA, USA) as a molecular weight marker. The UVP PhotoDoc-It Imaging System (Analytik, Jena, Germany) was used to view the end product and determine the genotype of each sample based on the number and size(s) of the DNA band(s) observed. Different banding patterns were indicative of different genotypes according to de Morais et al.
Confirmation of Genotyping Assay Results Using Sanger Sequencing
PCR products of both the CYP2C19*2 and CYP2C19*3 loci of all 22 pilot samples underwent Sanger sequencing. Eighteen PCR amplicons of the CYP2C19*2 locus from the main cohort underwent Sanger sequencing. These were systematically selected such that three samples, each representing a unique genotype and exhibiting acceptable DNA integrity, were selected from each PCR-restriction enzyme digestion run. 12 PCR amplicons of the CYP2C19*3 locus from the main cohort underwent Sanger sequencing; two from each PCR-restriction enzyme digestion run. All samples were sequenced by Sanger sequencing, at Macrogen Inc. (South Korea), using their Standard Bidirectional Sequencing service.
The resultant forward and reverse reads were analyzed using the BioEdit sequence alignment software (Informer Technologies Inc., Los Angeles, CA, USA) to confirm the genotype of each sample. For the CYP2C19*2 variant, the reads were simultaneously aligned against the dbSNP (National Center for Biotechnology Information, Bethesda, MD, USA) CYP2C19*2 reference sequence (rs4244285) to determine whether the 681G > A variant was absent or present on one or both alleles. For the CYP2C19*3 variant, the reads were simultaneously aligned against the dbSNP CYP2C19*3 reference sequence (rs4986893) to determine whether the 636G > A variant was absent or present.
Statistical Analysis
Chi-square analysis was used to evaluate the association between categorical variables. A P value of < 0.001 was deemed statistically significant. Odds ratios with 95% confidence intervals were calculated using a MedCalc Software Ltd. odds ratio calculator. https://www.medcalc.org/calc/odds_ratio.php (Version 22.009; accessed August 9, 2023).
Results
Pilot Cohort Genotype Frequencies, Demographics, and Clopidogrel Resistance
All 22 participants in the pilot cohort were genotyped for the CYP2C19*2 and the CYP2C19*3 variants. Genotypes obtained from the restriction digest protocols were fully concordant with those from Sanger sequencing. The CYP2C19*2 genotype, clopidogrel resistance phenotype, ethnicity and sex information are summarized in Table 1. Of the nine participants found to be phenotypically resistant, eight identified as Indian and one identified as African. The one African carried the heterozygous genotype and is male. The CYP2C19*3 was not found.
Cohort Demographic and Clinical Characteristics
A total of 162 participants were newly recruited for this study. All data including age, gender, ethnicity, medical history, medications, and genotypes of each participant are recorded in Supplementary Table 1. The participants ranged in age from 19 to 86 years with a mean age of 59 years and 82% (n = 133) of the cohort was ≥ 50 years. Male patients comprised 51.9% (n = 84) of the cohort while female patients comprised 48.1% (n = 78). The majority of participants self-identified as Indian at 65.4% (n = 106), while 22.8% (n = 37) self-identified as either Caribbean Black and 11.7% (n = 19) identified as mixed.
Coronary artery disease (CAD) (77.1%, n = 125), hypertension (HTN) (72.8%, n = 118), and type 2 diabetes mellitus (T2D) (46.9%, n = 76) were the most commonly diagnosed clinical conditions within the cohort. Clinical diagnosis of hypersensitivity lung disease (31.5%, n = 51), cardiovascular event (13%, n = 21), chronic kidney disease (4%, n = 7) and chronic obstructive pulmonary disease (2.5%, n = 4) were also reported in the cohort.
The relationship between disease prevalence and sex was determined using a chi-square test of independence. Prevalence of CAD was found to be significantly associated with sex in in this cohort, χ2(1, 162) = 11.8, p = 0.0006. There was no statistically significant association between sex and prevalence of HTN or T2D in this cohort, χ2(1, 162) = 0.18, p = 0.68 and χ2(1, 162) = 5.45, p = 0.02, respectively.
Overall, high rates of comorbidities among CAD, HTN and T2D were reported; 84% (105/125) of people presenting with CAD also presented with either T2D, HTN or both T2D & HTN. Strikingly, 45.6% (57/125) of people presenting with CAD also presented with both T2D and HTN. Of those participants presenting with T2D, 96.1% (73/76) also presented with either CAD, HTN, or both CAD & HTN.
In this cohort, the most common demographics were Indian males with CAD (35.2%, n = 57), Indian males with HTN (27.8%, n = 45), and Indian males with both CAD and HTN (25.9%, n = 42).
The most prescribed medications in the cohort at the time of data collection were aspirin (80.7%, n = 131), statins (70.4%, n = 114), ACEi/ARB (64.2%, n = 104), clopidogrel (42.0%, n = 68), and nitrates (34.0%, n = 55). Beta blockers (28.4%, n = 46), calcium channel blockers (27.2%, n = 44), trimetazidine (10.5%, n = 17), ivabradine (4.9%, n = 8) and mineralocorticoid receptor antagonist (4.3%, n = 7) were also prescribed to a lesser extent in the cohort.
In this cohort, aspirin plus statins was the most commonly prescribed combination of medications (64.8%, n = 105), followed by aspirin and ACEi/ARB (58.6%, n = 95), and aspirin and clopidogrel (38.9%, n = 63). Importantly, in this cohort, clopidogrel was never prescribed alone. It was most commonly prescribed with aspirin (92.6%, n = 63). It was also prescribed with statins and ACEi/ARB. In fact, clopidogrel was commonly prescribed in combination with aspirin, statins, and ACEi/ARB (64.7%, n = 44). Of those prescribed these four medications in combination, 56.8% (n = 25) were males of Indian descent. Of note, clopidogrel was almost twice as likely to be prescribed to males than females. This is in contrast to aspirin, statins, and ACEi/ARB, which were not significantly different in their rates of prescription to males and females.
CYP2C19*2 and CYP2C19*3 Genotype Frequencies
Of the newly recruited participants, 156 of 162 were successfully genotyped for the CYP2C19*2 (Table 2) and CYP2C19*3 variants. All genotyping results from the PCR and restriction digest protocols were consistent with the genotypes obtained from sequencing.
The CYP2C19*2 allele was detected in 63.5% (99/156) of the cohort. At the CYP2C19*2 locus, 57 participants had the GG (CYP2C19*1/*1) genotype, 65 had the AG (CYP2C19*1/*2) genotype and 34 had the AA (CYP2C19*2/*2) genotype.
The CYP2C19*3 variant was not detected in this study population. Visualization of restriction digested PCR amplicons from all samples showed only two DNA bands at 96 bp and 175 bp, indicating the genotype GG at the CYP2C19*3 locus.
The CYP2C19*2 allele was most prevalent in Indian patients, as 73% of Indians carried this allele. There was a statistically significant association between ethnicity and CYP2C19*2 genotype χ2(2, 156) = 18.449, p = 0.001. Compared to African patients, Indian patients were three times more likely (odds ratio (OR) = 3.18, 95% confidence interval (CI) 1.36–7.44) to have the AG genotype and four times more likely (OR = 4.31, 95% CI 1.93–9.6049) to have the AA or AG genotype. A chi-square test of independence showed that there was no statistically significant association between sex and CYP2C19*2 genotype, χ2(2, 156) = 2.39, p = 0.302.
CYP2C19*2 Genotype and Clopidogrel Usage in Patients with CAD
Of particular concern, almost two-thirds (63.6%; 42/66) of the patients prescribed clopidogrel carried the loss-of-function CYP2C19*2 variant (Table 3); three-quarters of which had the heterozygous CYP2C19*1/*2 genotype, and one-quarter had the homozygous CYP2C19*2/*2 genotype.
In this cohort derived from the major catchment clinic for patients with cardiovascular disease in this country, we found that 66.6% (80/120) of people presenting with CAD, i.e., those for whom clopidogrel is a common treatment option, carry the CYP2C19*2 loss-of-function allele; 53 of these had the heterozygous CYP2C19*1/*2 genotype, and 27 of these had the homozygous CYP2C19*2/*2 genotype. Even more strikingly, we found that 63.5% (40/63) of people with CAD who are already prescribed clopidogrel carry the CYP2C19*2 allele, three-quarters of which had the heterozygous CYP2C19*1/*2 genotype, and one-quarter had the homozygous CYP2C19*2/*2 genotype (Table 3).
CAD and T2D are common comorbidities in this cohort with 61 patients suffering from both. In this cohort, we found that a majority of 73.7% (45/61) of participants with both CAD and T2D carry the CYP2C19*2 allele; 18% (11/61) of which are homozygous for the CYP2C19*2 allele and 55.7% (34/61) of which are heterozygous for the CYP2C19*2 allele.
Discussion
We report here, for the first time in a Caribbean population, the allele and genotype frequencies of the two most common CYP2C19 loss-of-function alleles, CYP2C19*2 and CYP2C19*3, and clopidogrel usage in a clinically relevant CVD cohort in Trinidad. Trinidad has the highest reported prevalence of CVD in the Caribbean and clopidogrel is predominantly prescribed to lower the risk of adverse cardiovascular events. We found that the CYP2C19*2 allele, which is associated with clopidogrel resistance, is present at high rates in this population. In this study, 63.6% of the patients prescribed clopidogrel carried the CYP2C19*2 loss-of-function allele, and are therefore at heightened risk of MACE and other negative consequences of impaired CYP2C19 metabolic activation of clopidogrel. Our results indicate that the merits of CYP2C19 genotype-guided antiplatelet therapy to clinical practice in this country should be further investigated due to the substantive frequency of the CYP2C19*2 genotypes found in the CVD population of T&T.
In our pilot cohort, we investigated the relationship between CYP2C19 loss-of-function alleles and biochemically determined clopidogrel resistance. The results were as expected, in that the CYP2C19*2 genotype status was concordant with clopidogrel resistance, despite the small sample size. The one individual with the previously described poor metabolizer genotype (homozygous AA (CYP2C19*2/*2) genotype) showed clopidogrel resistance; 43% of the participants with the heterozygous AG (CYP2C19*1/*2) genotype showed clopidogrel resistance. This is expected as the one CYP2C19*2 allele should result in decreased drug metabolism in cases where the second allele does not have any additional effect on drug metabolism. However, we only genotyped for the CYP2C19*2 and CYP2C19*3 alleles and the nature of the second allele is not known in these cases. An important caveat for all genotyping assays is that any alleles not detected in the assay are designated as *1 including rare increased function, reduced function, or non-functional alleles that are not routinely screened for. Therefore, it is possible that in some cases, the second allele could compensate for the effect of the single CYP2C19*2 allele. Finally, two of the seven participants with the homozygous GG (CYP2C19*1/*1) genotype showed clopidogrel resistance while the rest showed normal metabolism. This may be explained by additional alleles that affect clopidogrel drug metabolism that were not tested for in this study such as variants in the ABCB1 gene.
Our main cohort, consisting of 162 patients with CVD collected from the main catchment clinic for the country, was balanced in terms of sex. Ethnic composition and clinical characteristics such as disease prevalence, comorbidities, and drug prescription trends were similar to previous reports for this population. CAD, HTN, and T2D were the most common clinically diagnosed conditions and comorbidities among these three were high. The most common demographics among CVD patients were Indian males with CAD, HTN, or both, consistent with previous reports. Clopidogrel was the most commonly prescribed antiplatelet therapy. Thus, despite the limited sample size, our cohort appears representative of this population.
The CYP2C19*2 variant was detected in 63.5% (99/156) of the cohort. This rate is two times greater than the previously reported rate of 37% for the Trinidadian population (Mungrue, Wyke, and Ramkissoon 2016). This discrepancy may be explained by the limited sample sizes of both studies. However, our cohort was derived from a clinically relevant population of patients with CVD who are either already being treated with or likely to be treated with clopidogrel whereas the previous study’s cohort was derived from the general population. We argue that the detection rate in our clinically relevant cohort is more pertinent to the use of clopidogrel as an antiplatelet therapy in this country. Given the high prevalence of cardiovascular mortality in this country [25], and the fact that clopidogrel is the most commonly prescribed antiplatelet therapy for the treatment of symptomatic CAD, this high detection rate of the CYP2C19*2 in the CVD population is of clinical concern.
In our cohort, 120 people presented with CAD. More than half of these (n = 63/120) are already prescribed clopidogrel and the rest may be prescribed clopidogrel in the future. Strikingly, we found that almost two-thirds (63.5%; 40/63) of people with CAD who are prescribed clopidogrel carry the CYP2C19*2 allele; ten homozygous and 30 heterozygous. Thus, in this cohort, the majority of patients with CAD currently being treated with clopidogrel are receiving a drug which they may be fully or partially resistant to. A further 17 CAD patients who were not prescribed clopidogrel at the time of data collection but may be in the future, also carried the CYP2C19*2 allele. The implications of these findings are worrying.
Adding to the complexity of the situation, there is a high rate of comorbidity of T2D and CAD in this population, and it is known that there is an increased rate of heart attack due to platelet aggregation in patients with T2D [23]. Consequently, efficacious antiplatelet therapy is particularly important in patients suffering from both CAD and T2D [24]. Yet, we found that 73.7% of participants with both CAD and T2D carry the CYP2C19*2 allele, and are therefore phenotypically likely poor or intermediate metabolizers of clopidogrel. This result indicates that the addition of CYP2C19 genotypic information for making drug treatment decisions for the very common class of individuals suffering from both CAD and T2D can have a substantive impact in this population. Moreover, the alternative drug ticagrelor has been shown to perform better than clopidogrel in diabetes mellitus patients with acute coronary syndrome (ACS) [28].
In our data, there was a statistically significant association between the Indian ethnicity and the CYP2C19*2 allele (χ2 = 17.67; df = 4; p = 0.001), in that, Indian patients are four times more likely (OR = 3.97, 95% CI 1.80–8.75) to carry this allele. This is consistent with previous international and local studies [9] (Mungrue, Wyke, and Ramkissoon (2016)). Also consistent with the previous studies, the CYP2C19*3 variant was undetected in this cohort, further emphasizing its rarity [5, 8, 32].
CYP2C19 genotyping prior to prescribing antiplatelet therapy in Trinidad has the potential to improve cardiovascular disease outcomes and lead to a more effective redistribution of government expenditure on the various antiplatelet options. In fact, implementing CYP2C19 genotype-guided therapy was found to be a cost-effective approach in guiding the selection of medication in patients with ACS undergoing percutaneous coronary intervention (PCI) [1]. In patients undergoing primary PCI in the POPular Genetics trial, a CYP2C19 genotype-guided strategy compared to universal ticagrelor or prasugrel treatment resulted in favorable cost-effectiveness with QALYs gained and cost savings [4]. We expect similar economic efficiencies to be gained in this population by genotype-guided strategies while improving clinical outcomes, particularly in the interest of South Asian patients with cardiovascular disease.
Inclusion of phenotypic clopidogrel-resistance data from the larger cohort would have helped to more robustly establish whether the association between CYP2C19*2 and CYP2C19*3 genetic variants and clopidogrel resistance holds in this population. However, this was not possible due to funding limitations. The findings of this study may be biased due to the small sample size. Further large-scale studies in this setting would be informative. Additionally, ancestry composition of the patients classified as mixed ethnicity was not calculated. This was beyond the scope of this study but could have facilitated identification of any genetic ancestry influence in detecting the CYP2C19*2 variant. Finally, only two variants were tested for. We appreciate that there may be other relevant variants at play in this population including rare and novel variants in this population. Nonetheless, this study provides sound evidence that CYP2C19 genotype is an important factor in the use of clopidogrel in this population.
Conclusions
We found a high prevalence of the CYP2C19*2 genetic variant, resulting in the substantial occurrence of poor and intermediate clopidogrel metabolizers in the CVD population of T&T. This is particularly true in the Indian sub-population , which is affected the most by CVD in this country. In a nation where CVD is the leading cause of all deaths, and clopidogrel is the most common antiplatelet therapy, the ubiquitous use of clopidogrel must be questioned. These data emphasize the clinical need for further investigation into whether CYP2C19 genotype-guided antiplatelet therapy, including the use of alternative drugs, should be applied to the CVD population of Trinidad and Tobago.
Data Availability
All data generated or analyzed during this study are included in this published article/as supplementary information files.
References
AlMukdad S, Elewa H, Al-Badriyeh D. Economic evaluations of CYP2C19 genotype-guided antiplatelet therapy compared to the universal use of antiplatelets in patients with acute coronary syndrome: a systematic review. J Cardiovasc Pharmacol Ther. 2020;25(3):201–11. https://doi.org/10.1177/1074248420902298. (PMID: 32027168).
Bapatla N, Ramoutar UD, Sharma N, Ramoutar A, Ortega VL, Goorachan A, Haffizulla F. Cardiovascular disease in the Indo-Caribbean population: a scoping review. Cureus. 2021;13(6): e15375.
Chadee D, Seemungal T, Pinto LM, Pereira MC, Maharaj R, Teelucksingh S. Prevalence of self-reported diabetes, hypertension and heart disease in individuals seeking state funding in Trinidad and Tobago, West Indies. J Epidemiol Global Health. 2013;3(2):95–103.
Claassens DMF, van Dorst PWM, Vos GJA, Bergmeijer TO, Hermanides RS, van’t Hof AWJ, van der Harst P, Barbato E, Morisco C, Tjon Joe Gin RM, Asselbergs FW, Mosterd A, Herrman JR, Dewilde WJM, Postma MJ, Deneer VHM, Ten Berg JM, Boersma C. Cost effectiveness of a CYP2C19 genotype-guided strategy in patients with acute myocardial infarction: results from the POPular genetics trial. Am J Cardiovasc Drugs. 2022;22(2):195–206. https://doi.org/10.1007/s40256-021-00496-4. (PMID: 34490590).
De Morais SM, Wilkinson GR, Blaisdell J, Meyer UA, Nakamura K, Goldstein JA. Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol Pharmacol. 1994;46(4):594–8.
Ellis KJ, Stouffer GA, McLeod HL, Lee CR. Clopidogrel pharmacogenomics and risk of inadequate platelet inhibition: US FDA recommendations. Pharmacogenomics. 2009;10(11):1799–817.
Fricke-Galindo I, Céspedes-Garro C, Rodrigues-Soares F, Naranjo MEG, Delgado Á, de Andrés F, López-López M, Peñas-Lledó E, LLerena A. Interethnic variation of CYP2C19 alleles, ‘predicted’ phenotypes and ‘measured’ metabolic phenotypes across world populations. Pharmacogenomics J. 2016. https://doi.org/10.1038/tpj.2015.70.
Ionova Y, Ashenhurst J, Zhan J, Nhan H, Kosinski C, Tamraz B, Chubb A. CYP2C19 Allele Frequencies in Over 2.2 Million Directto-Consumer Genetics Research Participants and the Potential Implication for Prescriptions in a Large Health System. Clin Transl Sci. 2020;13(6):1298–1306. https://doi.org/10.1111/cts.12830
Klein MD, Williams AK, Lee CR, Stouffer GA. Clinical utility of CYP2C19 genotyping to guide antiplatelet therapy in patients with an acute coronary syndrome or undergoing percutaneous coronary intervention. Arterioscler Thromb Vasc Biol. 2019;39(4):647–52.
Ko DT, Krumholz HM, Tu JV, Austin PC, Stukel TA, Koh M, Chong A, de Melo Jr JF, Jackevicius CA. Clinical outcomes of Plavix and generic clopidogrel for patients hospitalized with an acute coronary syndrome. Circ Cardiovasc Qual Outcomes. 2018;11(3): e004194.
Knuuti J, Wijns W, Saraste A, Capodanno D, Barbato E, Funck-Brentano C, Prescott E, Storey RF, Deaton C, Cuisset T, Agewall S, Dickstein K, Edvardsen T, Escaned J, Gersh BJ, Svitil P, Gilard M, Hasdai D, Hatala R, Mahfoud F, Masip J, Muneretto C, Valgimigli M, Achenbach S, Bax JJ, ESC Scientific Document Group. 2019 ESC guidelines for the diagnosis and management of chronic coronary syndromes. Eur Heart J. 2020;41(3):407–77. https://doi.org/10.1093/eurheartj/ehz425. (Erratum in: Eur Heart J. 2020 Nov 21;41(44):4242. PMID: 31504439).
Lee CR, Luzum JA, Sangkuhl K, Gammal RS, Sabatine MS, Stein CM, Kisor DF, et al. Clinical pharmacogenetics implementation consortium guideline for CYP2C19 genotype and clopidogrel therapy: 2022 update. Clin Pharmacol Ther. 2022;112(5):959–67.
Levine GN, Bates ER, Blankenship JC, Bailey SR, Bittl JA, Cercek B, Chambers CE, Ellis SG, Guyton RA, Hollenberg SM, Khot UN, Lange RA, Mauri L, Mehran R, Moussa ID, Mukherjee D, Nallamothu BK, Ting HH. 2011 ACCF/AHA/SCAI guideline for percutaneous coronary intervention: a report of the American college of cardiology foundation/American heart association task force on practice guidelines and the society for cardiovascular angiography and interventions. Circulation. 2011;124(23):e574-651. https://doi.org/10.1161/CIR.0b013e31823ba622. (Erratum in: Circulation. 2012 Feb 28; 125(8):e412. Dosage error in article text. PMID: 22064601).
Levine GN, Bates ER, Bittl JA, Brindis RG, Fihn SD, Fleisher LA, Granger CB, Lange RA, Mack MJ, Mauri L, Mehran R, Mukherjee D, Newby LK, O’Gara PT, Sabatine MS, Smith PK, Smith SC Jr. 2016 ACC/AHA guideline focused update on duration of dual antiplatelet therapy in patients with coronary artery disease: a report of the American college of cardiology/American heart association task force on clinical practice guidelines: an update of the 2011 ACCF/AHA/SCAI guideline for percutaneous coronary intervention, 2011 ACCF/AHA guideline for coronary artery bypass graft surgery, 2012 ACC/AHA/ACP/AATS/PCNA/SCAI/STS guideline for the diagnosis and management of patients with stable ischemic heart disease, 2013 ACCF/AHA guideline for the management of ST-elevation myocardial infarction, 2014 AHA/ACC guideline for the management of patients with non-ST-elevation acute coronary syndromes, and 2014 ACC/AHA guideline on perioperative cardiovascular evaluation and management of patients undergoing noncardiac surgery. Circulation. 2016;134(10):e123–55. https://doi.org/10.1161/CIR.0000000000000404.
Martin A, Downing J, Maden M, Fleeman N, Alfirevic A, Haycox A, Pirmohamed M. An assessment of the impact of pharmacogenomics on health disparities: a systematic literature review. Pharmacogenomics. 2017;18(16):1541–50.
Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, Walker JR, et al. Cytochrome P-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360(4):354–62.
de Morais SM, Wilkinson GR, Blaisdell J, Nakamura K, Meyer UA, Goldstein JA. The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J Biol Chem. 1994;269(22):15419–22.
Mungrue K, Wyke M, Ramkissoon V. The distribution of altered forms of CYP2C19* 2 allele in a Trinidadian population. Is clopidogrel the right choice. Eur J Bio Pharm Sci.
Park K-J, Chung H-S, Kim S-R, Kim H-J, Han J-Y, Lee S-Y. Clinical, pharmacokinetic, and pharmacogenetic determinants of clopidogrel resistance in Korean patients with acute coronary syndrome. Korean J Lab Med. 2011;31(2):91–4.
Patel AP, Wang M, Kartoun U, Ng K, Khera AV. Quantifying and understanding the higher risk of atherosclerotic cardiovascular disease among South Asian individuals: results from the UK biobank prospective cohort study. Circulation. 2021;144(6):410–22. https://doi.org/10.1161/CIRCULATIONAHA.120.052430.
Polasek TM, Doogue MP, Miners JO. Metabolic activation of clopidogrel. In vitro data provide conflicting evidence for the contributions of CYP2C19 and PON1. Ther Adv Drug Safety. 2011;2(6):253–61.
Razzaghi H, Martin DN, Quesnel-Crooks S, Hong Y, Gregg E, Andall-Brereton G, Gawryszweski V, Saraiya M. 10-year trends in noncommunicable disease mortality in the Caribbean region. Rev Panam Salud Publ Pan Am J Public Health. 2019;43: e37.
Schneider DJ. Factors contributing to increased platelet reactivity in people with diabetes. Diabetes Care. 2009;32(4):525–7.
Schuette C, Steffens D, Witkowski M, Stellbaum C, Bobbert P, Schultheiss H-P, Rauch U. The effect of clopidogrel on platelet activity in patients with and without type-2 diabetes mellitus: a comparative study. Cardiovasc Diabetol. 2015;14:15.
Seecheran NA, Maharaj A, Boodhai B, Seecheran R, Seecheran V, Persad S, Ramsaroop K, et al. Prevalence of clOpidogrel ‘resIstaNce’in a selected population of patients undergoing elective percutaneous coronary intervention at a tertiary cardiovascular centre in Trinidad: the POINT pilot study. Open Heart. 2019;6(1): e000841.
Shah KS, Patel J, Rifai MA, Agarwala A, Bhatt AB, Levitzky YS, Palaniappan L. Cardiovascular risk management in the south Asian patient: a review. Health Sci Rev (Oxf). 2022;4: 100045. https://doi.org/10.1016/j.hsr.2022.100045. (PMID: 36438886; PMCID: PMC9699691).
Sun B, Wen Y-F, Culhane-Pera KA, Lo M, Straka RJ. Pharmacogenomic variabilities in geo-ancestral subpopulations and their clinical implications: results of collaborations with Hmong in the United States. Front Genet. 2022;13:1070236.
Sweeny JM, Angiolillo DJ, Franchi F, Rollini F, Waksman R, Raveendran G, Dangas G, et al. Impact of diabetes mellitus on the pharmacodynamic effects of ticagrelor versus clopidogrel in troponin-negative acute coronary syndrome patients undergoing Ad hoc percutaneous coronary intervention. J Am Heart Assoc. 2017;6(4): e005650.
Valgimigli M, Bueno H, Byrne RA, Collet JP, Costa F, Jeppsson A, Jüni P, Kastrati A, Kolh P, Mauri L, Montalescot G, Neumann FJ, Petricevic M, Roffi M, Steg PG, Windecker S, Zamorano JL, Levine GN, ESC Scientific Document Group, ESC Committee for Practice Guidelines (CPG), ESC National Cardiac Societies. 2017 ESC focused update on dual antiplatelet therapy in coronary artery disease developed in collaboration with EACTS: the task force for dual antiplatelet therapy in coronary artery disease of the European society of cardiology (ESC) and of the European association for cardio-thoracic surgery (EACTS). Eur Heart J. 2018;39(3):213–60. https://doi.org/10.1093/eurheartj/ehx419. (PMID: 28886622).
Whirl-Carrillo M, Huddart R, Gong Li, Sangkuhl K, Thorn CF, Whaley R, Klein TE. An evidence-based framework for evaluating pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2021;110(3):563–72.
Yusuf S, Zhao F, Mehta SR, Chrolavicius S, Tognoni G, Fox KK, Clopidogrel in Unstable Angina to Prevent Recurrent Events Trial Investigators. Effects of clopidogrel in addition to aspirin in patients with acute coronary syndromes without ST-segment elevation. New Engl J Med. 2001;345(7):494–502.
Zand N, Tajik N, Moghaddam AS, Milanian I. Genetic polymorphisms of cytochrome P450 enzymes 2C9 and 2C19 in a healthy Iranian population. Clin Exp Pharmacol Physiol. 2007;34(1-2):102–5. https://doi.org/10.1111/j.1440-1681.2007.04538.x. (Erratum in: Clin Exp Pharmacol Physiol. 2007;34(4):385).
Zhou Y, Ingelman-Sundberg M, Lauschke VM. Worldwide distribution of cytochrome P450 alleles: a meta-analysis of population-scale sequencing projects. Clin Pharmacol Ther. 2017;102(4):688–700.
Acknowledgements
We thank the participants of the study.
Funding
No funding or sponsorship was received for this study or publication of this article. The Rapid Service Fee was funded by the authors.
Author information
Authors and Affiliations
Contributions
Daniele Jones: performed experiments, conducted analysis, wrote the manuscript. Shana Persad-Ramdeensingh: collected samples, performed experiments, conducted analysis, edited the manuscript. Sheherazade Crystal Abrahim; performed experiments. Naveen Seecheran: conceptualized the project, supervised the sample collection, edited the manuscript. Rajini Rani Haraksingh: conceptualized the project, supervised the experimental work and analysis, conducted analysis, wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors Daniele Jones, Shana Persad-Ramdeensingh, Sheherazade Crystal Abrahim, Naveen Seecheran, and Rajini Rani Haraksingh have nothing to disclose.
Ethical Approval
This study was performed in accordance with the Helsinki Declaration of 1964 and its later amendments. Ethical approval was obtained from the Campus Research Ethics Committee of The University of the West Indies, St. Augustine, Trinidad and Tobago (Reference Number CEC610/05/18). All participants were recruited under informed consent.
Additional information
Prior Presentation: Portions of this work have been previously presented orally at the following conferences: (1) Daniele Jones, Naveen Seecheran, Rajini Haraksingh. “Association of CYP2C19*2 and CYP2C19*3 allelic variants with clopidogrel resistance in cardiovascular disease patients in Trinidad, W.I.”. National Health Research Conference, Port of Spain, Trinidad & Tobago. 25th November 2022. (2) Daniele Jones, Shana Persad-Ramdeensingh, Naveen Seecheran, Rajini Haraksingh. “Evidence to Support CYP2C19 Genotype-guided Antiplatelet Therapy for Cardiovascular Disease Patients in Trinidad” National Health Research Conference Port of Spain, Trinidad & Tobago. 17th November 2023.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Jones, D., Persad-Ramdeensingh, S., Abrahim, S.C. et al. Prevalence of CYP2C19*2 and CYP2C19*3 Allelic Variants and Clopidogrel Use in Patients with Cardiovascular Disease in Trinidad & Tobago. Cardiol Ther 13, 191–203 (2024). https://doi.org/10.1007/s40119-024-00348-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40119-024-00348-7