Abstract
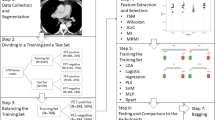
Our objective was to investigate radiomics signatures and prediction models defined by four segmentation methods in using 2-[18F]fluoro-2-deoxy-d-glucose positron emission tomography (18F-FDG PET) imaging of lung metastases of soft-tissue sarcomas (STSs). For this purpose, three fixed threshold methods using the standardized uptake value (SUV) and gradient-based edge detection (ED) were used for tumor delineation on the PET images of STSs. The Dice coefficients (DCs) of the segmentation methods were compared. The least absolute shrinkage and selection operator (LASSO) regression and Spearman’s rank, and Friedman’s ANOVA test were used for selection and validation of radiomics features. The developed radiomics models were assessed using ROC (receiver operating characteristics) curve and confusion matrices. According to the results, the DC values showed the biggest difference between SUV40% and other segmentation methods (DC: 0.55 and 0.59). Grey-level run-length matrix_run-length nonuniformity (GLRLM_RLNU) was a common radiomics signature extracted by all segmentation methods. The multivariable logistic regression of ED showed the highest area under the ROC (receiver operating characteristic) curve (AUC), sensitivity, specificity, and accuracy (AUC: 0.88, sensitivity: 0.85, specificity: 0.74, accuracy: 0.81). In our research, the ED method was able to derive a significant model of radiomics. GLRLM_RLNU which was selected from all segmented methods as a meaningful feature was considered the obvious radiomics feature associated with the heterogeneity and the aggressiveness. Our results have apparently showed that radiomics signatures have the potential to uncover tumor characteristics.




Similar content being viewed by others
References
K.G. Billingsley, M.E. Burt, E. Jara, R.J. Ginsberg, J.M. Woodruff, D.H.Y. Leung et al., Pulmonary metastases from soft tissue sarcoma: analysis of patterns of disease and postmetastasis survival. Ann. Surg. 229, 602 (1999). https://doi.org/10.1097/00000658-199905000-00002
G. Marulli, M. Mammana, G. Comacchio, F. Rea, Survival and prognostic factors following pulmonary metastasectomy for sarcoma. J. Thorac. Dis. 9, S1305 (2017)
M.F. Brennan, Soft tissue sarcoma: advances in understanding and management. Surgeon 3, 216 (2005). https://doi.org/10.1016/S1479-666X(05)80044-7
J.W. Fletcher, B. Djulbegovic, H.P. Soares, B.A. Siegel, V.J. Lowe, G.H. Lyman et al., Recommendations on the use of 18F-FDG PET in oncology. J. Nucl. Med. 49, 480 (2008). https://doi.org/10.2967/jnumed.107.047787
R.T. Hoppe, R.H. Advani, W.Z. Ai, R.F. Ambinder, P. Aoun, P. Armand et al., NCCN guidelines insights: hodgkin lymphoma, version 1.2018. J. Natl. Compr. Can Netw. 16, 245 (2018). https://doi.org/10.6004/jnccn.2018.0013
M. Vallières, C.R. Freeman, S.R. Skamene, I. El Naqa, A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys. Med. Biol. 60, 5471 (2015). https://doi.org/10.1088/0031-9155/60/14/5471
K.C. Genadry, S. Pietrobono, R. Rota, C.M. Linardic, Soft tissue sarcoma cancer stem cells: an overview. Front. Oncol. 8, 475 (2018). https://doi.org/10.3389/fonc.2018.00475
P. Lambin, R.G.P.M. Van Stiphout, M.H.W. Starmans, E. Rios-Velazquez, G. Nalbantov, H.J.W.L. Aerts et al., Predicting outcomes in radiation oncology-multifactorial decision support systems. Nat. Rev. Clin. Oncol. 10, 27 (2013). https://doi.org/10.1038/nrclinonc.2012.196
E. Scalco, G. Rizzo, Texture analysis of medical images for radiotherapy applications ELISA. BJR. 90, 20160642 (2017). https://doi.org/10.1259/bjr.20160642
B. Zhang, F. Ouyang, D. Gu, Y. Dong, L. Zhang, X. Mo et al., Advanced nasopharyngeal carcinoma: pre-treatment prediction of progression based on multi-parametric MRI radiomics. Oncotarget 8, 72457 (2017). https://doi.org/10.18632/oncotarget.19799
B. Foster, U. Bagci, A. Mansoor, Z. Xu, D.J. Mollura, A review on segmentation of positron emission tomography images. Comput Biol Med. 50, 76 (2014). https://doi.org/10.1016/j.compbiomed.2014.04.014
C.A. Owens, C.B. Peterson, C. Tang, E.J. Koay, W. Yu, D.S. Mackin et al., Lung tumor segmentation methods: impact on the uncertainty of radiomics features for non-small cell lung cancer. PLoS One 13, e0205003 (2018). https://doi.org/10.1371/journal.pone.0205003
M. Avanzo, J. Stancanello, I. El Naqa, Beyond imaging: the promise of radiomics. Phys. Medica. 38, 122 (2017). https://doi.org/10.1016/j.ejmp.2017.05.071
S. Ha, H. Choi, J.C. Paeng, G.J. Cheon, Radiomics in oncological PET/CT: a methodological overview. Nucl. Med. Mol. Imaging. 53, 14 (2019). https://doi.org/10.1007/s13139-019-00571-4
B.H. Byun, C.-B. Kong, J. Park, Y. Seo, I. Lim, C.W. Choi et al., Initial metabolic tumor volume measured by 18F-FDG PET/CT can predict the outcome of osteosarcoma of the extremities. J. Nucl. Med. 54, 1725 (2013). https://doi.org/10.2967/jnumed.112.117697
U. Nestle, S. Kremp, A. Schaefer-Schuler, C. Sebastian-Welsch, D. Hellwig, C. Rübe et al., Comparison of different methods for delineation of 18F-FDG PET-positive tissue for target volume definition in radiotherapy of patients with non-Small cell lung cancer. J. Nucl. Med. 46, 1342 (2005)
F. Orlhac, J.J.-A. Maisonobe, C.A. Garcia, B. Vanderlinden, M. Soussan, J.J.-A. Maisonobe et al., Tumor texture analysis in 18F-FDG PET: relationships between texture parameters, histogram indices, standardized uptake values, metabolic volumes, and total lesion glycolysis. J. Nucl. Med. 55, 414 (2014). https://doi.org/10.2967/jnumed.113.129858
M. Hatt, F. Tixier, L. Pierce, P.E. Kinahan, C. Cheze Le Rest, D. Visvikis et al., Characterization of PET/CT images using texture analysis: The past, the present… any future? Eur. J. Nucl. Med. Mol. Imaging. 44, 151 (2017). https://doi.org/10.1007/s00259-016-3427-0
P.E. Kinahan, J.W. Fletcher, Positron emission tomography-computed tomography standardized uptake values in clinical practice and assessing response to therapy. Semin Ultrasound CT MRI. 31, 496 (2010). https://doi.org/10.1053/j.sult.2010.10.001
J.A. Thie, Understanding the standardized uptake value, its methods, and implications for usage. J. Nucl. Med. 45, 1431 (2004)
M. Werner-Wasik, A.D. Nelson, W. Choi, Y. Arai, P.F. Faulhaber, P. Kang et al., What is the best way to contour lung tumors on PET scans? Multiobserver validation of a gradient-based method using a NSCLC digital PET phantom. Int. J. Radiat. Oncol. Biol. Phys. 82, 1164 (2012). https://doi.org/10.1016/j.ijrobp.2010.12.055
X. Geets, J.A. Lee, A. Bol, M. Lonneux, V. Grégoire, A gradient-based method for segmenting FDG-PET images: methodology and validation. Eur. J. Nucl. Med. Mol. Imaging. 32, 1427 (2007). https://doi.org/10.1007/s00259-006-0363-4
K. Clark, B. Vendt, K. Smith, J. Freymann, J. Kirby, P. Koppel et al., The cancer imaging archive (TCIA): maintaining and operating a public information repository. J. Digit. Imaging. 26, 1045 (2013). https://doi.org/10.1007/s10278-013-9622-7
Kelly H. Zou, Simon K. Warfield, Aditya Bharatha, et al. Statistical validation of image segmentation quality based on a spatial overlap index. Acad. Radiol. 11, 178 (2004). https://www.academicradiology.org/article/S1076-6332(03)00671-8/
C. Nioche, F. Orlhac, S. Boughdad, S. Reuze, J. Goya-Outi, C. Robert et al., Lifex: A freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res. 78, 4786 (2018). https://doi.org/10.1158/0008-5472.CAN-18-0125
F. Orlhac, M. Soussan, K. Chouahnia, E. Martinod, I. Buvat, 18F-FDG PET-derived textural indices reflect tissue-specific uptake pattern in non-small cell lung cancer. PLoS One 10, 1 (2015). https://doi.org/10.1371/journal.pone.0145063
R.M. Haralick, K. Shanmugam, I. Dinstein, Textural features for image classification. IEEE Trans. Syst. Man. Cybern. 3, 610 (2007). https://doi.org/10.1109/tsmc.1973.4309314
Xu D, Kurani AS, Furst JD, Raicu DS. Run-length Encoding for Volumetric Texture. In: 4th Iated Int Conf Vis Imaging, IMAGE Process (2004).
G. Thibault, B. Fertil, C. Navarro, S. Pereira, P. Cau, N. Levy et al., Texture indexes and gray level size zone matrix application to cell nuclei classification. Pattern Recognit. Inf. Process. (2009). https://doi.org/10.1142/S0218001413570024
H. Lee, D.S. Lee, H. Kang, B.N. Kim, M.K. Chung, Sparse brain network recovery under compressed sensing. IEEE Trans. Med. Imaging. 30, 1154 (2011). https://doi.org/10.1109/TMI.2011.2140380
W. Zhao, Y. Xu, Z. Yang, Y. Sun, C. Li, L. Jin et al., Development and validation of a radiomics nomogram for identifying invasiveness of pulmonary adenocarcinomas appearing as subcentimeter ground-glass opacity nodules. Eur. J. Radiol. 112, 161 (2019). https://doi.org/10.1016/j.ejrad.2019.01.021
R. Tibshirani, The lasso method for variable selection in the Cox model. Stat. Med. 16, 385 (1997)
M. Kirienko, L. Cozzi, L. Antunovic, L. Lozza, A. Fogliata, E. Voulaz et al., Prediction of disease-free survival by the PET/CT radiomic signature in non-small cell lung cancer patients undergoing surgery. Eur. J. Nucl. Med. Mol. Imaging. 45, 207 (2018). https://doi.org/10.1007/s00259-017-3837-7
Acknowledgements
Not applicable.
Funding
This work was supported by a Grant from the National Research Foundation of Korea (NRF) Grant funded by the Korea government (MSIT) (Nos. 2021R1F1A1050903, 2021M2E7A2079183) and the Korea Institute of Radiological and Medical Sciences (KIRAMS) funded by the Ministry of Science and ICT, Republic of Korea (50536–2021).
Author information
Authors and Affiliations
Contributions
HS, H-BS, and JYK contributed equally to this work. Writing—original draft: HS.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sheen, H., Shin, HB. & Kim, J.Y. Comparison of radiomics prediction models for lung metastases according to four semiautomatic segmentation methods in soft-tissue sarcomas of the extremities. J. Korean Phys. Soc. 80, 247–256 (2022). https://doi.org/10.1007/s40042-021-00360-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40042-021-00360-3