Abstract
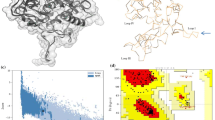
Camellia sinensis (L.) Kuntze (commonly called tea plant) is an economically important cash crop in India. This plant is prone to infection by several fungal pathogens. Chitinases (EC 3.2.1.14) are enzymes that play a significant role in plants by hydrolyzing the N-acetylglucosamine polymer chitin. The present study entails different in silico characterization of chitinase enzyme using complete chitinase cDNA sequence from C. sinensis (CsChi). Template crystal structure of class I chitinase from Oryza sativa (PDB ID: 2DKV) was used for homology modeling of the CsChi enzyme. The model structure was refined and verified by using Structural Analysis and Verification Server (SAVES). The predicted model was submitted to the Protein Model Data Base (PMDB ID: PM0079561) and was docked with the chitin ligand, obtained from ChemSpider database. The results revealed that Serine in the predicted active site at position 199 of the enzyme is responsible for strong hydrogen bonding affinity with the ligand. Results of the evolutionary analysis showed that the obtained amino acid sequence of the CsChi enzyme belongs to class Ib chitinase.




Similar content being viewed by others
References
Legrand M, Kauffmann S, Geoffroy P, Fritig B (1987) Biological function of pathogenesis-related proteins: four tobacco pathogenesis-related proteins are chitinases. Proc Natl Acad Sci USA 84:6750–6754
Roy SC, Chakraborty BN (2012) Analysis of chitinase gene specific transcript accumulation in tea [Camellia sinensis (L.) O. Kuntze] during induced systemic resistance by methyl jasmonate. Indian J Biotechnol 11:142–147
Wan J, Pentecost G (2013) Potential application of chitin signaling in engineering broad-spectrum disease resistance to fungal and bacterial pathogens in plants. Adv Crop Sci Technol 1:1000e113
Sharma N, Sharma KP, Gaur RK, Gupta VK (2011) Role of chitinase in plant defense. Asian J Biochem 6:29–37
Grover A (2012) Plant chitinases: genetic diversity and physiological roles. Crit Rev Plant Sci 31:57–73
Kasprzewska A (2003) Plant chitinases—regulation and function. Cell Mol Biol Lett 8:809–824
Collinge DB, Kragh KM, Mikkelsen JD, Nielsen KK, Rasmussen U, Vad K (1993) Plant chitinases. Plant J 3:31–40
de A Gerhardt LB, Sachetto-Martins G, Contarini MG, Sandroni M, de P Ferreira R, de Lima VM, Cordeiro MC, de Oliveira DE, Margis-Pinheiro M (1997) Arabidopsis thaliana class IV chitinase is early induced during the interaction with Xanthomonas campestris. FEBS Lett 419:69–75
Chandra S, Chakraborty N, Chatterjee A, Rai R, Bera B, Acharya K (2014) Abiotic elicitor-mediated improvement of innate immunity in Camellia sinensis. J Plant Growth Regul 33(4):849–859
Nisha SN, Prabu GR, Mandal AKA, Arvinth S (2011) Purification and molecular characterization of chitinase from tea leaves [Camellia sinensis (L.) O. Kuntze]. Two A Bud 58:44–47
Saravanakumara D, Vijayakumar C, Kumar N, Samiyappan R (2007) PGPR-induced defense responses in the tea plant against blister blight disease. Crop Prot 26:556–565
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from trans-membrane regions. Nat Methods 8:785–786
Guruprasad K, Reddy BVP, Pandit MW (1990) Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Prot Eng 4:155–164
Ikai AJ (1980) Thermo stability and aliphatic index of globular proteins. J Biochem 88:1895–1898
Kyte J, Doolottle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In: Walker JM (ed) The proteomics protocols handbook. Humana Press, Totowa, pp 571–607
Garnier J, Gibrat JF, Robson B (1996) GOR secondary structure prediction method version IV. Methods Enzymol 266:540–553
Guermeur Y (1997) Combinaison de classifieurs statistiques, Application a la prediction de structure secondaire des proteins. Ph. D. Thesis, Université Paris
Geourjon C, Deleage G (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci 11:681–684
Garnier J, Gibrat JF, Robson B (1996) GOR secondary structure prediction method version IV. Methods Enzymol 266:540–553
Levin JM, Robson B, Garnier J (1986) An algorithm for secondary structure determination in proteins based on sequence similarity. FEBS Lett 205(2):303–308
Geourjon C, Deleage G (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci 11(6):681–684
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling. Bioinformatics 22:195–201
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Chandra S, Chatterjee S, Bandyopadhyay SK, Acharya K (2014) In silico molecular modeling and structural analysis of peroxidase enzymes from five different plants species. Res J Pharm Biol Chem Sci 5:1416–1427
Bowie JU, Lüthy R, Eisenberg D (1991) A method to identify protein sequences that fold into a known three-dimensional structure. Science 253(5016):164–170
Lüthy R, Bowie JU, Eisenberg D (1992) Assessment of protein models with three-dimensional profiles. Nature 356(6364):83–85
Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucl Acids Res 34:W116–W118
Duhovny D, Nussinov R, Wolfson HJ (2002) Efficient unbound docking of rigid molecules. In: Gusfield et al. (ed) Proceedings of the 2nd Workshop on Algorithms in Bioinformatics(WABI) Rome, Lecture Notes in Computer Science 2452, Springer Verlag, Italy, pp 185–200
Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ (2005) PatchDock and SymmDock: servers for rigid and symmetric docking. Nucl Acids Res 33:363–367
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res 25:4876–4882
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Brogue K, Chet I, Holliday M, Cressman R, Biddle P, Knowlton S, Mauvais CJ, Broglie R (1991) Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 254:1194–1197
Chatterjee S, Laskar A, Chatterjee A, Mandal C, Chaudhuri S (2011) In silico structural analysis of an immunotherapeutic glycoprotein T11TS (sheep CD58). Int J Biol Med Res 2:346–359
Sarma K, Dehury B, Sahu J, Sarmah R, Sahoo S, Sahu M, Sen P, Modi MK, Barooah M (2012) A comparative proteomic approach to analyse structure, function and evolution of rice chitinases: a step towards increasing plant fungal resistance. J Mol Model 18:4761–4780
Roy SC (2014) Molecular cloning and expression of tea chitinase gene in Pichia pastoris. Int J Adv Biotechnol Res 5(4):612–618
Mayer RT, McCollum TG, Niedz RP, Hearn CJ, McDonald RE, Berdis E, Doostdar H (1996) Characterization of seven basic endochitinases isolated from cell culture of Citrus sinensis (L.). Planta 200:289–295
Wang S, Shao B, Fu H, Rao P (2009) Isolation of a thermostable legumes chitinase and study on the antifungal activity. Appl Microbiol Biotechnol 85:313–321
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132
George RA, Heringa J (2003) An analysis of protein domain linkers: their classification and role in protein folding. Protein Eng 15(11):871–879
Gokhale RS, Khosla C (2000) Role of linkers in communication between protein modules. Curr Opin Chem Biol 4(1):22–27
Eisenberg D, Lüthy R, Bowie JU (1997) VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol 277:396–404
Yang AY, Källblad P, Mancera RL (2004) Molecular modeling prediction of ligand binding site flexibility. J Comput Aided Mol Des 18:235–250
Takaya N, Yamazaki D, Horiuchi H, Ohta A, Takagi M (1998) Cloning and characterization of a chitinase-encoding gene (ChiA) from Aspergillus nidulans, disruption of that decreases germination frequency and hyphal growth. Biosci Biotechnol Biochem 62:60–65
Hamid R, Khan MA, Ahmad M, Ahmad MM, Abdin MZ, Musarrat J, Javed S (2013) Chitinases: an update. J Pharm BioAllied Sci 5(1):21–29
Kuranda MJ, Robbins PW (1991) Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J Biol Chem 266:19758–19767
Durand A, Hughes R, Roussel A, Flatman R, Henrissat B, Juge N (2005) Emergence of a subfamily of xylanase inhibitors within glycoside hydrolase family 18. FEBS J 272(7):1745–1755
Funkhouser JD, Aronson NN (2007) Chitinase family GH18: evolutionary insights from the genomic history of a diverse protein family. BMC Evol Biol 7:96
Santos P, Fortunato A, Ribeiro A, Pawlowski K (2008) Chitinases in root nodules. Plant Biotechnol 25:299–307
Tyler L, Bragg JN, Wu J, Yang X, Tuskan GA, Vogel JP (2010) Annotation and comparative analysis of the glycoside hydrolase genes in Brachypodium distachyon. BMC Genomics 11:600
Wiweger M, Farbos I, Ingouff M, Lagercrantz U, von Arnold S (2003) Expression of Chia4-Pa chitinase genes during somatic and zygotic embryo development in Norway spruce (Picea abies): similarities and differences between gymnosperm and angiosperm class IV chitinases. J Exp Bot 54:2691–2699
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chandra, S., Dutta, A.K., Chandrashekara, K.N. et al. In silico characterization, homology modeling of Camellia sinensis chitinase and its evolutionary analyses with other plant chitinases. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 87, 685–695 (2017). https://doi.org/10.1007/s40011-015-0634-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-015-0634-6