Abstract
Purpose
Thorough knowledge of the nature and frequency of co-infections is essential to optimize treatment strategies and risk assessment in cases of coronavirus disease 2019 (COVID-19). This study aimed to evaluate the multiplex polymerase chain reaction (PCR) screening approach for community-acquired bacterial pathogens (CABPs) at hospital admission, which could facilitate identification of bacterial co-infections in hospitalized COVID-19 patients.
Methods
Clinical data and biomaterials from 200 hospitalized COVID-19 patients from the observational cohort of the Competence Network for community-acquired pneumonia (CAPNETZ) prospectively recruited between March 17, 2020, and March 12, 2021 in 12 centers in Germany and Switzerland, were included in this study. Nasopharyngeal swab samples were analyzed on hospital admission using multiplex real-time reverse transcription (RT)-PCR for a broad range of CABPs.
Results
In total of 200 patients Staphylococcus aureus (27.0%), Haemophilus influenzae (13.5%), Streptococcus pneumoniae (5.5%), Moraxella catarrhalis (2.5%), and Legionella pneumophila (1.5%) were the most frequently detected bacterial pathogens. PCR detection of bacterial pathogens correlated with purulent sputum, and showed no correlation with ICU admission, mortality, and inflammation markers. Although patients who received antimicrobial treatment were more often admitted to the ICU and had a higher mortality rate, PCR pathogen detection was not significantly related to antimicrobial treatment.
Conclusion
General CABP screening using multiplex PCR with nasopharyngeal swabs may not facilitate prediction or identification of bacterial co-infections in the early phase of COVID-19-related hospitalization. Most patients with positive PCR results appear to be colonized rather than infected at that time, questioning the value of routine antibiotic treatment on admission in COVID-19 patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Coronavirus disease 2019 (COVID-19), which is caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), emerged in China in late 2019 [1] and subsequently led to a worldwide pandemic. Although most cases of COVID-19 are characterized by mild or uncomplicated respiratory illnesses, patients may show complications such as co-infections, acute respiratory distress syndrome, and sepsis [2, 3]. Co-infections frequently occur in patients with respiratory viral illnesses such as influenza, and current guidelines recommend empirical antibiotic treatment and investigations for co-infections in patients with a severe clinical course [4,5,6]. For COVID-19, detailed data on community-acquired bacterial co-infections is scarce, which could be attributed to the reduced sampling frequency to reduce the risk of exposure for health care workers collecting and processing respiratory samples [7]. A co-infection can be defined as an infection occurring concurrently with the initial SARS-CoV-2 infection, while super-infections occur during the clinical course of the disease. However, these two entities are often not clearly defined in the literature, leading to varying published rates of co- and/or super-infections in patients with COVID-19.
The most frequently identified pathogens in CAP with co-infections are respiratory syncytial virus, metapneumovirus, influenza virus, Staphylococcus aureus, Streptococcus pneumoniae, Haemophilus influenzae, Mycoplasma pneumoniae, and Legionella pneumophila [8,9,10,11,12,13]. Previous reports have identified high rates of empirical antibiotic use, contrasting the low rates of laboratory-confirmed co-infections in COVID-19 [14,15,16]. For example, in one study, confirmed community-onset bacterial co-infection was reported in only 3.5% of the cases (of which 1.8% were bloodstream infections and 1.7% involved respiratory pathogens), although more than 50% of the patients were treated with empirical antibiotic therapy [17]. Overuse of antimicrobials increases the risk of adverse effects such as multidrug-resistant nosocomial secondary infections [18]. Thus, knowledge of the nature and frequency of co-infections is warranted to optimize antibiotic use in line with antibiotic stewardship principles (ABS).
Multiplex polymerase chain reaction (PCR) panels can rapidly identify the presence of relevant respiratory pathogens and may help clarify the indications for antimicrobial use as well as the choice of drugs. Therefore, this systematic prospective observational multicenter study aimed to determine whether general multiplex PCR-based screening for community-acquired bacterial pathogens (CABPs) at hospital admission could help physicians identify bacterial co-infections and predict the clinical course of COVID-19.
Methods
Study population
Clinical data and biological specimens from 200 patients with COVID-19 between March 17, 2020, and March 12, 2021 included in the German multicenter observational competence network for community-acquired pneumonia (CAPNETZ) [19] were evaluated in this study. 200 patients were recruited from various CAPNETZ study centers in Switzerland and in Germany: Berlin Charité Campus Benjamin Franklin (n = 45), Dortmund (n = 44), Frankfurt (n = 34), Lübeck (n = 32), Munich (n = 21), Bad Arolsen (n = 9), Gerlingen (n = 8), Rotenburg (n = 3), Basel (n = 2), Hannover (n = 1) and Bonn (n = 1). Every patient underwent a standardized medical assessment at the first presentation to the participating site, which included documentation of comorbidities, medical therapy, and antibiotics, details on inpatient hospital treatment, vaccination status, diagnostic procedures, laboratory parameters, and clinical symptoms. Routine microbiological work-up on admission included blood and sputum culture, pneumococcal and legionella urinary antigen. Follow-up evaluations involved assessment of structured outcome parameters at days 3, 7, and 28 after study inclusion.
Inclusion and exclusion criteria*
The following inclusion criteria were defined:
-
Age ≥ 18 years.
-
Presentation of a new pulmonary infiltrate on chest radiography or computed tomography (CT).
-
Positive results on SARS-CoV-2 PCR testing.
-
Presence of at least one of the following clinical symptoms: cough, purulent sputum, positive auscultation, or fever.
Patients were excluded from the CAPNETZ study cohort if they met any of the following criteria:
-
Newly diagnosed active tuberculosis within the last 2 months.
-
Hospitalization > 48 h prior to inclusion.
-
Noninfectious pulmonary infiltrates.
*inclusion/exclusion criteria were modified during the study period by a protocol amendment of CAPNETZ study group. Main reason for the protocol amendment was to facilitate the inclusion of patients with COVID-19. From the 13th of October 2020 onwards, only the criteria in bold needed to be evaluated for inclusion or exclusion of patients. 43 of 200 patients were included after the amendment of the study protocol. All patients presented clinical symptoms. 5 of 43 patients had been already hospitalized > 48 h prior to inclusion. We decided not to exclude these patients from the analysis, because SARS-CoV-2 infections were clearly community-acquired. Chest radiography/ computed tomography (CT) was either not performed (n = 9) or showed no pulmonary infiltrates (n = 19) in 28 of 43 patients.
Sample collection
Nasopharyngeal swabs and blood samples were collected according to a standardized protocol within 48 h after hospital admission. To guarantee optimal pathogen detection quality, a standardized swab-collection and transport system for respiratory bacteria (nylon-flocked swab for nasopharyngeal collection, Product No. 503CS01, Hain Lifescience) in accordance with the CLSI standard M40-A for quality control of specimen transport devices was used. In addition, blood samples were obtained for routine clinicochemical diagnostics.
Laboratory testing
Respiratory sample processing was performed at two clinical laboratory sites (Institute of Virology, University of Freiburg; Department of Infectious Diseases and Microbiology, Lübeck, Germany) accredited according to DIN EN ISO 15189. Samples were analyzed using a multiplex real-time RT-PCR panel previously described by Bierbaum et al. [20] to test for respiratory viruses, including adenovirus, bocavirus, coronavirus (CoV) OC43, CoV 229E, CoV HKU1, CoV NL63, enterovirus, influenza virus A + B, human metapneumonvirus, parainfluenza virus 1–4, human parechovirus, respiratory syncytial virus A + B, and rhinovirus. In addition, all samples were tested for CABPs, including Staphylococcus aureus, Haemophilus influenzae, Streptococcus pneumoniae, Moraxella catarrhalis, Bordetella pertussis, Legionella pneumophila, Chlamydophila pneumoniae, and Mycoplasma pneumoniae (ampliCube respiratory bacterial panel, Fa. Mikrogen). Laboratory confirmation of SARS-CoV-2 was performed at local centers. Blood samples were processed at the local study centers.
Ethics statement
This study was approved by the Institutional Ethics Committee of the Hannover Medical School (Project approval number: 301-2008) and reviewed by the Institutional Ethics Committees (IECs) of all participating institutions, which operate according to the Declaration of Helsinki. Written consent was obtained from all the participants.
Definitions and statistical analyses
Continuous data were described as median (25th and 75th percentiles) and categorical data were presented as absolute and relative frequencies. Relevant characteristics of the groups were compared using the Mann–Whitney U test (continuous variables) and chi-square test (categorical variables). Statistical hypothesis testing was performed using two-sided exploratory significance levels of 0.05*. Statistical analyses were performed using Microsoft Excel 2013 and IBM SPSS Statistics version 25.0 (IBM Corp, Armonk, NY). The number of patients included in the analysis was predetermined by the study protocol.
Results
Baseline characteristics
A total of 200 patients with confirmed SARS-CoV-2 infection were included; the median age was 58.5 years, and 63.5% of the patients were male. Cough and fever were the most frequent clinical symptoms and median oxygen saturation at admission was 91.7%. Relevant laboratory findings on admission are presented in Supplementary Table 1. While 48.0% of the patients had a preexisting cardiovascular comorbidity, 16.0% had a preexisting pulmonary comorbidity, 19.0% had diabetes mellitus, and 7.0% had a preexisting malignancy. Of these, 23.4% were admitted to intensive care units (ICUs), and of those 15.2% required mechanical ventilation. Median duration of hospital stay was 9 days and at a follow-up assessment after 3 days, 83.5% were still in hospital care, while 81.5% were discharged home 28 days after study inclusion. The 30-day mortality rate was 4.5%. Empirical antibiotic therapy was initiated in 51.5%, with ampicillin/sulbactam, ceftriaxone and piperacillin/tazobactam as the most relevant drugs. Median duration of antibiotic treatment was 5 days. In 38.8% of the patients, a combination of a beta-lactam antibiotic and a macrolide antibiotic was administered. All relevant baseline characteristics are shown in Supplementary Table 1.
Frequency and spectrum of CABPs
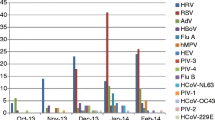
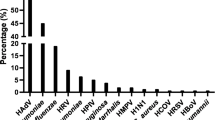
In total, 43.0% of the study patients showed positive CABP PCR screening results with at least one bacterial pathogen. Of those, 17.4% presented with more than one simultaneous microbial pathogen in addition to SARS-CoV-2. Staphylococcus aureus (27.0%) was the predominant bacterial pathogen, followed by Haemophilus influenzae (13.5%), Streptococcus pneumoniae (5.5%), Moraxella catarrhalis (2.5%), and Legionella pneumophila (1.5%). Mycoplasma pneumoniae, Chlamydophila pneumoniae, and Bordetella pertussis were not detected. Findings of routine microbiological work-up on admission are illustrated in Supplementary Table 1. Positive viral PCR screening results were obtained in 1.0% of the patients, and the only detectable viral pathogen was rhinovirus.
Comparison of COVID-19 patients with and without positive CAPB PCR screening results
Table 1 shows a comparison of patients with and without positive CAPB PCR screening results. The two groups showed no significant differences in vital signs, duration of hospital stay, levels of inflammatory markers (C-reactive protein [CRP], procalcitonin [PCT], and leucocytes), ICU admission rates, and mortality rates. Purulent sputum and higher platelet counts were associated with positive CABP PCR results. Table 2 shows a subgroup analysis of the clinical outcomes and inflammation parameters of COVID-19 patients with positive CABP PCR screening results in relation to the use of antimicrobial treatment. Patients with a positive CABP PCR screening who received antimicrobial treatment did not show better clinical outcomes for ICU admission and mortality. The duration of hospital stay was significantly longer in the group receiving antimicrobial treatment (11 days vs. 7 days; p < 0.01), while inflammation markers (CRP, p = 0.43; PCT, p = 0.06) did not significantly differ between the two groups (Table 2).
Discussion
A better understanding of the epidemiology of bacterial co-infections in COVID-19 patients is essential for risk stratification, therapeutic strategies, and infection surveillance. Identification of the causative pathogens of potential secondary infections and the use of optimal targeted antimicrobial therapy in line with ABS principles are essential in this regard. We used a cohort of well-characterized patients with laboratory-confirmed SARS-CoV-2 infection to evaluate the value of multiplex screening PCR for CABPs in identifying bacterial co-infections in COVID-19 patients. In the current study, 43.0% of the patients showed positive CAPB PCR screening results. Other studies have reported lower bacterial pathogen yields ranging from 28.0 to 32.0% [21, 22]. We attribute the higher incidence of bacterial pathogens in the present study to the following reasons: the routine use of culture-based microbiological diagnostics from deep respiratory samples is usually restricted to ICU patients. Consequently, most of the previous COVID-19 studies analyzed ICU patients, whereas most of the patients in the current study (76.4%) were admitted in the normal wards.
The low frequency and low yield of standard culture-based microbiological diagnostics in COVID-19 patients that are not intubated and suffer from only non-productive cough raises the question of how community-acquired co-infection in the early phase of the disease can be reliably detected [16]. To address this question, a multiplex PCR panel was used instead of a culture-based microbiological approach for pathogen detection in our study. However, a major drawback of this multiplex PCR approach is the difficulty in differentiating between colonization and infection. This particularly applies to nasopharyngeal swabs, which are usually used in normal ward patients. Up to 30.0% of the human population consists of asymptomatic nasal carriers of S. aureus [23]. S. pneumoniae nasal colonization is present in 6.5% of healthy adults [24], and H. influenzae can be found in the nasopharynx of 20.0% of healthy adults [25]. Considering the similarities between these high colonization rates and the detection rates of the predominant pathogens in the present study (S. aureus: 27.0%, S. pneumoniae: 5.5%, H. influenzae: 13.5%), a high proportion of positive PCR findings obtained from nasopharyngeal swabs in this study may represent colonization rather than infection. Autopsy studies [26] have reported that potential bacterial lung super-infections occur in approximately 32% of severe COVID-19 cases. In contrast, the incidence of bacterial lung co-infection in the early phase of hospitalization is expected to be lower. Hughes et al. [15] investigated the incidence of bacterial and fungal co-infections in hospitalized COVID-19 patients and found a low frequency of bacterial co-infections in the early COVID-19 hospital presentation. Positive blood cultures were identified in 7.1% (60/836) of the patients, of which 65% were classified as contaminants. Bacteremia caused by respiratory infection was confirmed in only two cases. As demonstrated in Table 2, COVID-19 patients with positive results in CABP PCR screening who received antimicrobial treatment did not show better clinical outcomes (regarding ICU admission and mortality) than patients with positive CABP PCR findings and no antimicrobial treatment. Furthermore, the duration of hospital stay of COVID-19 patients who showed a positive CABP PCR screening result and received antimicrobial treatment was significantly longer than that of COVID-19 patients who showed positive CABP PCR results and did not receive antimicrobial therapy (11 vs. 7 days, p = 0.003). In summary, these findings suggest that positive CABP PCR results are not correlated with the presence of bacterial co-infections in COVID-19. Consequently, clinical correlation of positive PCR findings by using adjunctive clinical and laboratory markers is needed to determine the significance of detected pathogens. In the current study, purulent sputum and platelet counts were correlated with positive CABP PCR screening results. Nevertheless, ideal clinical or laboratory parameters to distinguish pulmonary infection from colonization still do not exist. In this context, procalcitonin (PCT) seems to be the most reliable marker [27], although PCT can neither rule out nor confirm an ongoing bacterial infection. In our study, PCT levels were not significantly higher in the positive-PCR group than in the negative-PCR group, which might be related to the immunomodulatory treatment of COVID-19 patients. Giacobbe et al. [28] compared CRP and PCT values in COVID-19 patients with bloodstream infections treated with and without immunomodulatory therapy. The median CRP and PCT values of patients receiving neither tocilizumab nor steroids were higher than those of patients treated with steroids or tocilizumab.
The multicenter design of the study adds further significance to our findings and emphasizes the importance of well-characterized cohorts and biobanks. Nevertheless, the study has several limitations: we did neither analyze asymptomatic SARS-CoV-2-negative controls nor asymptomatic SARS-CoV-2-positive controls. Compared to other COVID-19 trials, mortality rate of the current study was quite low which might be related to the young age and the low ICU admission rate of the study cohort. Due to the prospective study design, patient had to give their written consent for inclusion into the study. Furthermore, the sample size was limited; therefore, analyses of larger cohorts in further studies are warranted to clarify the value of CABP PCR screening in COVID-19.
Conclusion
In our cohort of 200 COVID-19 patients, 43% showed a positive CABP PCR screening result within 48 h after hospital admission. Positive CABP PCR results were not associated with higher ICU or mortality rates, even in patients who did not receive antimicrobial treatment. Thus, a large proportion of CABPs detected using PCR on hospital admission may not cause secondary pulmonary infection during hospitalization. Discrimination between infection and colonization in PCR-positive patients on the basis of clinical signs or inflammation markers is difficult, particularly for S. aureus. Therefore, multiplex PCR findings obtained from nasopharyngeal swabs may not be a useful tool for streamlining antimicrobial treatment for COVID-19 patients in the early phase of hospitalization.
Availability of data and materials
All relevant data are made available in the manuscript and supplementary files.
References
Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–33.
Richardson S, Hirsch JS, Narasimhan M, Crawford JM, McGinn T, Davidson KW, et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA. 2020;323:2052–9.
Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–20.
Uyeki TM, Bernstein HH, Bradley JS, Englund JA, File TM, Fry AM, et al. Clinical practice guidelines by the Infectious Diseases Society of America: 2018 update on diagnosis, treatment, chemoprophylaxis, and institutional outbreak management of seasonal Influenzaa. Clin Infect Dis. 2019;68:e1–47.
Ewig S, Kolditz M, Pletz M, Altiner A, Albrich W, Dromann D, et al. Management of Adult Community-Acquired Pneumonia and Prevention - Update 2021 - Guideline of the German Respiratory Society (DGP), the Paul-Ehrlich-Society for Chemotherapy (PEG), the German Society for Infectious Diseases (DGI), the German Society of Medical Intensive Care and Emergency Medicine (DGIIN), the German Viological Society (DGV), the Competence Network CAPNETZ, the German College of General Practitioneers and Family Physicians (DEGAM), the German Society for Geriatric Medicine (DGG), the German Palliative Society (DGP), the Austrian Society of Pneumology Society (OGP), the Austrian Society for Infectious and Tropical Diseases (OGIT), the Swiss Respirator6.y Society (SGP) and the Swiss Society for Infectious Diseases Society (SSI)]. Pneumologie. 2021. (Epub ahead of print). 2004;32(4):234–8
von Baum H, Schweiger B, Welte T, Marre R, Suttorp N, Pletz MW, et al. How deadly is seasonal influenza-associated pneumonia? The German Competence Network for Community-Acquired Pneumonia. Eur Respir J. 2011;37:1151–7.
Zhao S, Perry MR, Kennedy S, Wilson J, Chase-Topping ME, Anderson E et al. Risk factors for carbapenemase-producing organisms among inpatients in Scotland: a national matched case-control study. Infect Control Hosp Epidemiol. 2020;26(12):1–10
Feldman C, Anderson R. The role of co-infections and secondary infections in patients with COVID-19. Pneumonia (Nathan). 2021;13:5.
Langford BJ, So M, Raybardhan S, Leung V, Westwood D, MacFadden DR, et al. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect. 2020;26:1622–9.
Lai CC, Wang CY, Hsueh PR. Co-infections among patients with COVID-19: the need for combination therapy with non-anti-SARS-CoV-2 agents? J Microbiol Immunol Infect. 2020;53:505–12.
Lansbury L, Lim B, Baskaran V, Lim WS. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect. 2020;81:266–75.
Wee LE, Ko KKK, Ho WQ, Kwek GTC, Tan TT, Wijaya L. Community-acquired viral respiratory infections amongst hospitalized inpatients during a COVID-19 outbreak in Singapore: co-infection and clinical outcomes. J Clin Virol. 2020;128:104436.
Loubet P, Mathieu P, Lenzi N, Galtier F, Lainé F, Lesieur Z, et al. Characteristics of human Metapneumovirus infection in adults hospitalized for community-acquired influenza-like illness in France, 2012–2018: a retrospective observational study. Clin Microbiol Infect. 2021;27:127.e1-6.e6.
Furukawa D, Graber CJ. Antimicrobial stewardship in a pandemic: picking up the pieces. Clin Infect Dis. 2021;72:e542–4.
Hughes S, Troise O, Donaldson H, Mughal N, Moore LSP. Bacterial and fungal coinfection among hospitalized patients with COVID-19: a retrospective cohort study in a UK secondary-care setting. Clin Microbiol Infect. 2020;26:1395–9.
Karami Z, Knoop BT, Dofferhoff ASM, Blaauw MJT, Janssen NA, van Apeldoorn M, et al. Few bacterial co-infections but frequent empiric antibiotic use in the early phase of hospitalized patients with COVID-19: results from a multicentre retrospective cohort study in the Netherlands. Infect Dis (Lond). 2021;53:102–10.
Vaughn VM, Gandhi T, Petty LA, Patel PK, Prescott HC, Malani AN, et al. Empiric antibacterial therapy and community-onset bacterial co-infection in patients hospitalized with COVID-19: A multi-hospital cohort study. Clin Infect Dis. 2020; pp 1–2
Sticchi C, Alberti M, Artioli S, Assensi M, Baldelli I, Battistini A, et al. Regional point prevalence study of healthcare-associated infections and antimicrobial use in acute care hospitals in Liguria. Italy J Hosp Infect. 2018;99:8–16.
Welte T, Suttorp N, Marre R. CAPNETZ-community-acquired pneumonia competence network. Infection. 2004;32:234–8.
Bierbaum S, Forster J, Berner R, Rücker G, Rohde G, Neumann-Haefelin D, et al. Detection of respiratory viruses using a multiplex real-time PCR assay in Germany, 2009/10. Arch Virol. 2014;159:669–76.
Contou D, Claudinon A, Pajot O, Micaëlo M, Longuet Flandre P, Dubert M, et al. Bacterial and viral co-infections in patients with severe SARS-CoV-2 pneumonia admitted to a French ICU. Ann Intensive Care. 2020;10:119.
Baskaran V, Lawrence H, Lansbury LE, Webb K, Safavi S, Zainuddin NI, et al. Co-infection in critically ill patients with COVID-19: an observational cohort study from England. J Med Microbiol. 2021;70.
Sakr A, Brégeon F, Mège JL, Rolain JM, Blin O. Staphylococcus aureus Nasal colonization: an update on mechanisms, epidemiology, risk factors, and subsequent infections. Front Microbiol. 2018;9:2419.
Adler H, Nikolaou E, Gould K, Hinds J, Collins AM, Connor V, et al. Pneumococcal colonization in healthy adult research participants in the conjugate vaccine era, United Kingdom, 2010–2017. J Infect Dis. 2019;219:1989–93.
Slack MPE. A review of the role of Haemophilus influenzae in community-acquired pneumonia. Pneumonia (Nathan). 2015;6:26–43.
Clancy CJ, Schwartz IS, Kula B, Nguyen MH. Bacterial superinfections among persons with coronavirus Disease 2019: A comprehensive review of data from postmortem studies. Open Forum Infect Dis. 2021;8:ofab065.
Holm A, Pedersen SS, Nexoe J, Obel N, Nielsen LP, Koldkjaer O, et al. Procalcitonin versus C-reactive protein for predicting pneumonia in adults with lower respiratory tract infection in primary care. Br J Gen Pract. 2007;57:555–60.
Giacobbe DR, Battaglini D, Ball L, Brunetti I, Bruzzone B, Codda G, et al. Bloodstream infections in critically ill patients with COVID-19. Eur J Clin Investig. 2020;50:e13319.
Acknowledgements
Members of the CAPNETZ study group: M. Dreher, C. Cornelissen, W. Knüppel, D. Stolz, N. Suttorp, P. Creutz, A. Mikolajewska, A. le Claire, M. Benzke, T. Bauer, D. Krieger, M. Prediger, S. Schmager, M. Kolditz, B. Schulte-Hubbert, S. Langner, O. Degen, A. Hüfner, C. Hoffmann, T. Welte, J. Freise, G. Barten-Neiner, M. Nawrocki, I. Fuge, J. Naim, W. Kröner, T. Illig, N. Klopp, C. Kroegel, A. Moeser, B. Schleenvoigt, C. Forstner, D. Drömann, P. Parschke, K. Franzen, J. Rupp, N. Käding, E. Wouters, K. Walraven, D. Braeken, C. Spinner, H. Buschmann, A. Zaruchas, T. Schaberg, I. Hering, W. Albrich, F. Waldeck, F. Rassouli, S. Baldesberger, M. Panning, M. Wallner. The investigators of this scientific work acknowledge CAPNETZ STIFTUNG and the competence network CAPNETZ for project support with regard to using biomaterials and clinical data. CAPNETZ is a multidisciplinary approach to better understand and treat patients with CAP. The network has been made possible by the contributions of many investigators. Financial support. CAPNETZ is supported by the German Center for Lung Research (DZL) FKZ: 82DZL002A4.
Funding
Open Access funding enabled and organized by Projekt DEAL. Christoph Spinner reports grants, personal fees, and non-financial support from AbbVie; grants, personal fees, and non-financial support from Apeiron; grants, personal fees from Braun; grants from Cepheid; personal fees from Formycon; grants, personal fees, and non-financial support from Gilead Sciences; grants and personal fees from Eli Lilly; grants, personal fees, and non-financial support from Janssen-Cilag; grants, personal fees, and non-financial support from GSK/ViiV Healthcare; grants, personal fees, and non-financial support from MSD, outside the submitted work. Mathias W. Pletz reports grants from Pfizer and personal fees from MSD, Thermofisher, Ciesi, Novartis, GSK and Bayer. Jochen Schneider reports grants, personal fees, and non-financial support from AbbVie, Gilead, Janssen and Dr. Falk GmbH.
Author information
Authors and Affiliations
Consortia
Contributions
KR, CDS, and JS conceived the study. KR, JS, MP, MWP, ML, GR, JR, MW, JE, AE, MCSG contributed to acquisition of the data. KR, JS, FE, CDS analysed the data and interpreted the results. KR, FE performed statistical calculations. KR and JS wrote the manuscript. KR, JS, MP, MWP, ML, GR, JR, MW, JE, AE, FE, MCSG revised it critically for important intellectual content. All authors agree with the article submission. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no potential conflicts of interest.
Consent to participate
Signed informed consent was obtained from every individual for prospective biobanking.
Ethics approval
The study received approval from a central institutional review board. Approval of the Institutional Review Board of each center was also obtained. The study was carried out following definitions of Good Clinical Practice, according to the declaration of Helsinki.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rothe, K., Spinner, C.D., Panning, M. et al. Evaluation of a multiplex PCR screening approach to identify community-acquired bacterial co-infections in COVID-19: a multicenter prospective cohort study of the German competence network of community-acquired pneumonia (CAPNETZ). Infection 49, 1299–1306 (2021). https://doi.org/10.1007/s15010-021-01720-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s15010-021-01720-8