Abstract
Purpose
Methicillin-resistant Staphylococcus aureus (MRSA) is defined as S. aureus genetically having the mecA or mecC genes or phenotypically showing minimum inhibitory concentration (MIC) of oxacillin higher than 2 mg/L. However, recently, cefoxitin/oxacillin-susceptible mecA-positive S. aureus (OS-MRSA) has been reported worldwide. Little is known about the prevalence and virulence of these strains among clinically significant isolates in the UK. The aims were to (1) investigate the prevalence of OS-MRSA in seven major hospitals in the Wessex region/UK from a cohort of 500 clinically significant phenotypically identified MSSA isolates, (2) genetically characterise OS-MRSA strains by pulsed-field gel electrophoresis (PFGE) and compare these to common UK epidemic strains; and (3) to determine Panton-Valentine leukocidin (PVL; lukFS) gene carriage rates among these isolates.
Results
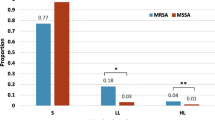
OS-MRSA was found in six isolates (1.2 %) of phenotypically identified and reported MSSA isolates by conventional methods. PFGE showed OS-MRSA strains to be genetically diverse and distinct from the common UK epidemic strains EMRSA-15 and EMRSA-16. None of these OS-MRSA stains carried the genes encoding PVL; however, overall positivity rate for PVL was 4.4 %, much higher than the nationally reported rates of 2 % in the UK.
Conclusion
There are still many unknowns regarding phenotypic and/or genetic characterization of the emerging OS-MRSA isolates in the UK and worldwide. Data regarding their epidemiology and optimal therapy for infection are limited and need further investigation not only in the UK, but also worldwide, as it is likely to have an impact on the empirical treatment of S. aureus infections.


Similar content being viewed by others
References
Tomassz A, Nachman S, Leaf H. Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob Agents Chemother. 1991;35:124–9.
Rohrer S, Maki H, Berger-Bächi B. What makes resistance to methicillin heterogeneous? J Med Microbiol. 2003;52:605–7.
Saeed K, Marsh P, Ahmad N. Cryptic resistance in Staphylococcus aureus: a risk for the treatment of skin infection? Curr Opin Infect Dis. 2014;27:130–6.
Saeed K, Dryden M, Parnaby R. Oxacillin-susceptible MRSA, the emerging MRSA clone in the UK? J Hosp Infect. 2010;76:267–8.
Saeed K, Green S, Ahmed N, Dryden M. oxacillin-susceptible methicillin-resistant Staphylococcus aureus: an emerging threat for UK hospitals? Poster presentation No 16 at the Health Protection Agency annual Conference, Warwick/UK. September 2011. https://www.hpa-events.org.uk/hpa/frontend/reg/absViewDocumentFE.csp?popup=1&documentID=4047&eventID=127. Accessed 01 April 2014.
Cuirolo A, Canigia LF, Gardella N, et al. Oxacillin- and cefoxitin-susceptible methicillin-resistant Staphylococcus. Int J Antimicrob Agents. 2011;37:178–9.
Kumar VA, Steffy K, Chatterjee M, et al. Detection of oxacillin-susceptible mecA-positive Staphylococcus aureus isolates using chromogenic medium MRSA ID. J Clin Microbiol. 2013;51:318–9.
Chambers HF. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin Microbiol Rev. 1997;10:781–91.
Hososaka Y, Hanaki H, Endo H, et al. Characterization of oxacillin-susceptible mecA-positive Staphylococcus aureus: a new type of MRSA. J Infect Chemother. 2007;13:79–86.
He W, Chen H, Zhao C, Zhang F, Wang H. Prevalence and molecular typing of oxacillin-susceptible mecA-positive Staphylococcus aureus from multiple hospitals in China. Diagn Microbiol Infect Dis. 2013;77:267–9.
Edward JP, Cartwrighta BC, Gavin K, et al. Use of Vitek 2 antimicrobial susceptibility profile to identify mecC in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2013;51:2732–4.
García-Garrote F, Cercenado E, Marín M, et al. Methicillin-resistant Staphylococcus aureus carrying the mecC gene: emergence in Spain and report of a fatal case of bacteraemia. J Antimicrob Chemother. 2014;69:45–50.
Cuny C, Layer F, Strommenger B, Witte W. Rare occurrence of methicillin-resistant Staphylococcus aureus CC130 with a novel mecA homologue in humans in Germany. PLoS One. 2011;6:e24360.
Ito T, Hiramatsu K, Tomasz A, et al. Guidelines for reporting novel mecA gene homologues. Antimicrob Agents Chemother. 2012;56:4997–9.
García-Álvarez L, Holden MTG, Lindsay H, et al. Methicillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11:595–603.
Public Health England, UK Standards for Microbiology Investigations. http://www.hpa.org.uk/ProductsServices/MicrobiologyPathology/UKStandardsForMicrobiologyInvestigations/TermsOfUseForSMIs/AccessToUKSMIs/. Accessed on 31 Jan 2014.
BSAC methods for antimicrobial susceptibility testing version 11.1 May 2012. http://www.bsac.org.uk. Accessed on 31 Jan 2014.
McDonald RR, Antonishyn NA, Hansen T, et al. Development of a triplex real-time PCR assay for detection of Panton-Valentine leukocidin toxin genes in clinical isolates of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:6147–9.
Lina G, Pie´mont Y, Godail-Gamot F, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–32.
Bruno Pichon, Robert Hill, Frederic Laurent, et al. Development of a real-time quadruplex PCR assay for simultaneous detection of nuc, Panton–Valentine leucocidin (PVL), mecA and homologue mecALGA251. J Antimicrob Chemother 2012; 67: 2338–2341 doi:10.1093/jac/dks221. Accessed 11 June 2012.
Murchan S, Kaufmann ME, Deplano A, et al. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J Clin Microbiol. 2003;41:1574–85.
Paterson GK, Larsen AR, Robb A, et al. The newly described mecA homologue, mecA(LGA251), is present in methicillin-resistant Staphylococcus aureus isolates from a diverse range of host species. J Antimicrob Chemother. 2012;67:2809–13.
Shore AC, Deasy EC, Slickers P, et al. Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55:3765–73.
Laurent F, Chardon H, Haenni M, et al. MRSA harboring mecA variant gene mecC, France. Emerg Infect Dis. 2012;18:1465–7.
Hellman J, Olsson-Liljequist B. A report on Swedish antibiotic utilisation and resistance in human medicine. Swedish Institute for Communicable Disease Control, Solna, Sweden. 2012. www.smi.se/publikationer or www.sva.se. Accessed 12 Oct 2013.
Schaumburg F, Köck R, Mellmann A, et al. Population dynamics among methicillin-resistant Staphylococcus aureus isolates in Germany during a 6-year period. J Clin Microbiol. 2012;50:3186–92.
Basset P, Prod’hom G, Senn L, et al. Very low prevalence of methicillin-resistant Staphylococcus aureus carrying the mecC gene in western Switzerland. J Hosp Infect. 2013;83:257–9.
Medhus A, Slettemeas JS, Marstein L, et al. Methicillin-resistant Staphylococcus aureus with the novel mecC gene variant isolated from a cat suffering from chronic conjunctivitis. J Antimicrob Chemother. 2013;68:968–9.
Paterson GK, Morgan FJ, Harrison EM, et al. Prevalence and characterization of human mecC methicillin-resistant Staphylococcus aureus isolates in England. J Antimicrob Chemother. 2014;69:907–10.
Holmes A, Ganner M, McGuane S, et al. Staphylococcus aureus isolates carrying Panton-Valentine leucocidin genes in England and Wales: frequency, characterization, and association with clinical disease. J Clin Microbiol. 2005;43:2384–90.
Pu W, Su Y, Li J, Li C, Yang Z, et al. High Incidence of oxacillin-susceptible mecA-Positive Staphylococcus aureus (OS-MRSA) Associated with Bovine Mastitis in China. PLoS One. 2014;9:e88134. doi:10.1371/journal.pone.0088134.
Giannouli S, Labrou M, Kyritsis A, et al. Detection of mutations in the FemXAB protein family in oxacillin-susceptible mecA-positive Staphylococcus aureus clinical isolates. J Antimicrob Chemother. 2010;65:626–33.
Ikonomidis A, Michail G, Vasdeki A, et al. In vitro and in vivo evaluations of oxacillin efficiency against mecA-positive oxacillin-susceptible Staphylococcus aureus. Antimicrob Agents Chemother. 2008;52:3905–8.
Labrou M, Michail G, Ntokou E, et al. Activity of οxacillin versus vancomycin against oxacillin-susceptible mecA-Positive Staphylococcus aureus clinical isolates by population analyses, time-kill assays and a murine thigh infection model. Antimicrob Agents Chemother. 2012;56:3388–91.
Sakoulas G, Gold HS, Venkataraman L, et al. Methicillin-resistant Staphylococcus aureus: comparison of susceptibility testing methods and analysis of mecA-positive susceptible strains. J Clin Microbiol. 2001;39:3946–51.
Acknowledgments
We thank staff in the microbiology departments of the participating hospitals for their support. This study was supported by ASPIRE, an independent research program funded by Pfizer.
Conflict of interest
There are no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Saeed, K., Ahmad, N., Dryden, M. et al. Oxacillin-susceptible methicillin-resistant Staphylococcus aureus (OS-MRSA), a hidden resistant mechanism among clinically significant isolates in the Wessex region/UK. Infection 42, 843–847 (2014). https://doi.org/10.1007/s15010-014-0641-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s15010-014-0641-1