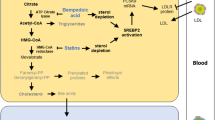
Abstract
Background
The use of circulating lipid traits as biomarkers to predict the risk of amyotrophic lateral sclerosis (ALS) is currently controversial, and the evidence-based medical evidence for the use of lipid-lowering agents, especially statins, on ALS risk remains insufficient. Our aim was to apply a Mendelian randomization (MR) approach to assess the causal impact of lipid-lowering agents and circulating lipid traits on ALS risk.
Materials and methods
Our study included primary and secondary analyses, in which the risk associations of lipid-lowering gene inhibitors, lipid traits, and ALS were assessed by the inverse variance weighting method as the primary approach. The robustness of the results was assessed using LDSC assessment, conventional MR sensitivity analysis, and used Mediating MR to explore potential mechanisms of occurrence. In the secondary analysis, the association of lipid-lowering genes with ALS was validated using the Summary data-based Mendelian Randomization (SMR) method.
Results
Our results showed strong evidence between genetic proxies for Apolipoprotein B (ApoB) inhibitor (OR = 0.76, 95% CI = 0.68 − 0.86; P = 5.58 × 10−6) and reduced risk of ALS. Additionally, 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR) inhibitor (OR = 1.06, 95% CI = 0. 85–1.33) was not found to increase ALS risk. SMR results suggested that ApoB expression was associated with increased ALS risk, and colocalization analysis did not support a significant common genetic variation between ApoB and ALS. Mediator MR analysis suggested a possible mediating role for interleukin-6 and low-density lipoprotein cholesterol (LDL-C). While elevated LDL-C was significantly associated with increased risk of ALS among lipid traits, total cholesterol (TC) and ApoB were weakly associated with ALS. LDSC results suggested a potential genetic correlation between these lipid traits and ALS.
Conclusions
Using ApoB inhibitor can lower the risk of ALS, statins do not trigger ALS, and LDL-C, TC, and ApoB levels can predict the risk of ALS.



Similar content being viewed by others
Data availability
Circulating lipid traits are available at http://csg.sph.umich.edu/willer/public/lipids2013/ (Used to obtain HDL-C, LDL-C, TC and TG). https://www.ebi.ac.uk/gwas/publications/27005778 (Used to obtain ApoB and ApoA1). Aggregated data for ALS and IL-6 are available from the GWAS Catalog: https://www.ebi.ac.uk/gwas/publications/29566793; https://www.ebi.ac.uk/gwas/publications/27989323. CAD: http://www.cardiogramplusc4d.org/data-downloads/. eQTL data for lipid-lowering drug targets gene were obtained from the GTEx V8 (https://gtexportal.org/) and eQTLGen (https://www.eqtlgen.org/) Consortium.
Abbreviations
- MR:
-
Mendelian Randomization
- SMR:
-
Summary data-based Mendelian Randomization
- HMGCR:
-
3-Hydroxy-3-methylglutaryl-coenzyme A reductase
- NPC1L1:
-
Niemann–Pick C1-Like 1
- PCSK9:
-
Proprotein convertase subtilisin/kexin type 9
- CETP:
-
Cholesteryl Ester Transfer Protein
- LDL-C:
-
Low-density lipoprotein cholesterol
- HDL-C:
-
High-density lipoprotein cholesterol
- TC:
-
Total cholesterol
- TG:
-
Triglycerides
- ApoB:
-
Apolipoprotein B
- ApoA1:
-
Apolipoprotein A1
- ALS:
-
Amyotrophic lateral sclerosis
- CAD:
-
Coronary artery disease
- IL-6:
-
Interleukin-6
- IV:
-
Instrumental variable
- OR:
-
Odds ratio
- CI:
-
Confidence interval
- IVW:
-
Inverse variance weighting method
- GLGC:
-
Global Lipid Genetics Consortium
- GWAS:
-
Genome-wide association study
- NCBI:
-
National Center for Biotechnology Information
- CARDIoGRAM:
-
Coronary ARtery DIsease Genome wide Replication and Meta-analysis
- LD:
-
Linkage disequilibrium
References
Marcus R (2022) What is amyotrophic lateral sclerosis? JAMA 328(24):2466. https://doi.org/10.1001/jama.2022.19305
Oskarsson B, Gendron TF, Staff NP (2018) Amyotrophic lateral sclerosis: an update for 2018. Mayo Clin Proc 93(11):1617–1628. https://doi.org/10.1016/j.mayocp.2018.04.007
Cutler RG, Pedersen WA, Camandola S, Rothstein JD, Mattson MP (2002) Evidence that accumulation of ceramides and cholesterol esters mediates oxidative stress-induced death of motor neurons in amyotrophic lateral sclerosis. Ann Neurol 52(4):448–457. https://doi.org/10.1002/ana.10312
Diekstra FP, Saris CG, van Rheenen W, Franke L, Jansen RC, van Es MA et al (2012) Mapping of gene expression reveals CYP27A1 as a susceptibility gene for sporadic ALS. PLoS ONE 7(4):e35333. https://doi.org/10.1371/journal.pone.0035333
Golomb BA, Kwon EK, Koperski S, Evans MA (2009) Amyotrophic lateral sclerosis-like conditions in possible association with cholesterol-lowering drugs: an analysis of patient reports to the University of California, San Diego (UCSD) Statin Effects Study. Drug Saf 32(8):649–661. https://doi.org/10.2165/00002018-200932080-00004
Liu J, Luo X, Chen X, Shang H (2020) Lipid profile in patients with amyotrophic lateral sclerosis: a systematic review and meta-analysis. Front Neurol 11:567753. https://doi.org/10.3389/fneur.2020.567753
Chio A, Calvo A, Ilardi A, Cavallo E, Moglia C, Mutani R et al (2009) Lower serum lipid levels are related to respiratory impairment in patients with ALS. Neurology 73(20):1681–1685. https://doi.org/10.1212/WNL.0b013e3181c1df1e
Kioumourtzoglou MA, Seals RM, Gredal O, Mittleman MA, Hansen J, Weisskopf MG (2016) Cardiovascular disease and diagnosis of amyotrophic lateral sclerosis: a population based study. Amyotroph Lateral Scler Frontotemporal Degener 17(7–8):548–554. https://doi.org/10.1080/21678421.2016.1208247
Weisskopf MG, Levy J, Dickerson AS, Paganoni S, Leventer-Roberts M (2022) Statin medications and amyotrophic lateral sclerosis incidence and mortality. Am J Epidemiol 191(7):1248–1257. https://doi.org/10.1093/aje/kwac054
Thompson PD, Panza G, Zaleski A, Taylor B (2016) Statin-associated side effects. J Am Coll Cardiol 67(20):2395–2410. https://doi.org/10.1016/j.jacc.2016.02.071
Golomb BA, Verden A, Messner AK, Koslik HJ, Hoffman KB (2018) Amyotrophic lateral sclerosis associated with statin use: a disproportionality analysis of the fda’s adverse event reporting system. Drug Saf 41(4):403–413. https://doi.org/10.1007/s40264-017-0620-4
Hoffman KB, Kraus C, Dimbil M, Golomb BA (2012) A survey of the FDA’s AERS database regarding muscle and tendon adverse events linked to the statin drug class. PLoS ONE 7(8):e42866. https://doi.org/10.1371/journal.pone.0042866
Schmidt AF, Finan C, Gordillo-Maranon M, Asselbergs FW, Freitag DF, Patel RS et al (2020) Genetic drug target validation using Mendelian randomisation. Nat Commun 11(1):3255. https://doi.org/10.1038/s41467-020-16969-0
Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S et al (2013) Discovery and refinement of loci associated with lipid levels. Nat Genet 45(11):1274–1283. https://doi.org/10.1038/ng.2797
Kettunen J, Demirkan A, Wurtz P, Draisma HH, Haller T, Rawal R et al (2016) Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat Commun 7:11122. https://doi.org/10.1038/ncomms11122
Nicolas A, Kenna KP, Renton AE, Ticozzi N, Faghri F, Chia R et al (2018) Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron 97(6):1268–83. https://doi.org/10.1016/j.neuron.2018.02.027
Brooks BR (1994) El escorial world federation of neurology criteria for the diagnosis of amyotrophic lateral sclerosis. subcommittee on motor neuron diseases/amyotrophic lateral sclerosis of the world federation of neurology research group on neuromuscular diseases and the EL escorial “clinical limits of amyotrophic lateral sclerosis” workshop contributors. J Neurol Sci 124:96–107. https://doi.org/10.1016/0022-510x(94)90191-0
Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S et al (2015) A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 47(10):1121–1130. https://doi.org/10.1038/ng.3396
Burgess S, Thompson SG, Collaboration CCG (2011) Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol 40(3):755–764. https://doi.org/10.1093/ije/dyr036
Shim H, Chasman DI, Smith JD, Mora S, Ridker PM, Nickerson DA et al (2015) A multivariate genome-wide association analysis of 10 LDL subfractions, and their response to statin treatment, in 1868 Caucasians. PLoS ONE 10(4):e0120758. https://doi.org/10.1371/journal.pone.0120758
Xu S, Wang P, Fung WK, Liu Z (2022) A novel penalized inverse-variance weighted estimator for Mendelian randomization with applications to COVID-19 outcomes. Biometrics. https://doi.org/10.1111/biom.13732
Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE et al (2016) Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet 48(5):481–487. https://doi.org/10.1038/ng.3538
Bowden J, Davey Smith G, Burgess S (2015) Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 44(2):512–525. https://doi.org/10.1093/ije/dyv080
Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics C et al (2015) LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47(3):291–5. https://doi.org/10.1038/ng.3211
Chauquet S, Zhu Z, O’Donovan MC, Walters JTR, Wray NR, Shah S (2021) Association of antihypertensive drug target genes with psychiatric disorders: a mendelian randomization study. JAMA Psychiat 78(6):623–631. https://doi.org/10.1001/jamapsychiatry.2021.0005
Wallace C (2013) Statistical testing of shared genetic control for potentially related traits. Genet Epidemiol 37(8):802–813. https://doi.org/10.1002/gepi.21765
Plagnol V, Smyth DJ, Todd JA, Clayton DG (2009) Statistical independence of the colocalized association signals for type 1 diabetes and RPS26 gene expression on chromosome 12q13. Biostatistics 10(2):327–334. https://doi.org/10.1093/biostatistics/kxn039
Lu CH, Allen K, Oei F, Leoni E, Kuhle J, Tree T et al (2016) Systemic inflammatory response and neuromuscular involvement in amyotrophic lateral sclerosis. Neurol Neuroimmunol Neuroinflamm 3(4):e244. https://doi.org/10.1212/NXI.0000000000000244
Ahola-Olli AV, Wurtz P, Havulinna AS, Aalto K, Pitkanen N, Lehtimaki T et al (2017) Genome-wide association study identifies 27 loci influencing concentrations of circulating cytokines and growth factors. Am J Hum Genet 100(1):40–50. https://doi.org/10.1016/j.ajhg.2016.11.007
Woolf WB, Zagkos L, Gill D (2022) TwoStepCisMR: a novel method and r package for attenuating bias in cis-mendelian randomization analyses. Genes (Basel). https://doi.org/10.3390/genes13091541
Sanderson E. Multivariable Mendelian Randomization and Mediation (2021) Cold Spring Harb Perspect Med 11(2). doi: https://doi.org/10.1101/cshperspect.a038984.
Elliott DA, Weickert CS, Garner B (2010) Apolipoproteins in the brain: implications for neurological and psychiatric disorders. Clin Lipidol 51(4):555–573. https://doi.org/10.2217/CLP.10.37
Nishimura AL, Mitne-Neto M, Silva HC, Richieri-Costa A, Middleton S, Cascio D et al (2004) A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet 75(5):822–831. https://doi.org/10.1086/425287
Ingre C, Chen L, Zhan Y, Termorshuizen J, Yin L, Fang F (2020) Lipids, apolipoproteins, and prognosis of amyotrophic lateral sclerosis. Neurology 94(17):e1835–e1844. https://doi.org/10.1212/WNL.0000000000009322
Kim SM, Noh MY, Kim H, Cheon SY, Lee KM, Lee J et al (2017) 25-Hydroxycholesterol is involved in the pathogenesis of amyotrophic lateral sclerosis. Oncotarget 8(7):11855–11867. https://doi.org/10.18632/oncotarget.14416
Schaper F, Rose-John S (2015) Interleukin-6: Biology, signaling and strategies of blockade. Cytokine Growth Factor Rev 26(5):475–487. https://doi.org/10.1016/j.cytogfr.2015.07.004
Campbell IL, Erta M, Lim SL, Frausto R, May U, Rose-John S et al (2014) Trans-signaling is a dominant mechanism for the pathogenic actions of interleukin-6 in the brain. J Neurosci 34(7):2503–2513. https://doi.org/10.1523/JNEUROSCI.2830-13.2014
Pronto-Laborinho A, Pinto S, Gromicho M, Pereira M, Swash M, de Carvalho M (2019) Interleukin-6 and amyotrophic lateral sclerosis. J Neurol Sci 398:50–53. https://doi.org/10.1016/j.jns.2019.01.026
Garbuzova-Davis S, Ehrhart J, Sanberg PR, Borlongan CV (2018) Potential role of humoral IL-6 cytokine in mediating pro-inflammatory endothelial cell response in amyotrophic lateral sclerosis. Int J Mol Sci. https://doi.org/10.3390/ijms19020423
Saeedi Saravi SS, Saeedi Saravi SS, Arefidoust A, Dehpour AR (2017) The beneficial effects of HMG-CoA reductase inhibitors in the processes of neurodegeneration. Metab Brain Dis 32(4):949–965. https://doi.org/10.1007/s11011-017-0021-5
Rosoff DB, Bell AS, Jung J, Wagner J, Mavromatis LA, Lohoff FW (2022) Mendelian randomization study of pcsk9 and hmg-coa reductase inhibition and cognitive function. J Am Coll Cardiol 80(7):653–662. https://doi.org/10.1016/j.jacc.2022.05.041
Nabizadeh F, Balabandian M, Sharafi AM, Ghaderi A, Rostami MR, Naser Moghadasi A (2022) Statins and risk of amyotrophic lateral sclerosis: a systematic review and meta-analysis. Acta Neurol Belg 122(4):979–986. https://doi.org/10.1007/s13760-021-01753-8
Zheng Z, Sheng L, Shang H (2013) Statins and amyotrophic lateral sclerosis: a systematic review and meta-analysis. Amyotroph Lateral Scler Frontotemporal Degener 14(4):241–245. https://doi.org/10.3109/21678421.2012.732078
Acknowledgements
The authors thank all the participants and researchers who contributed and collected data.
Funding
This research was supported by the National Natural Science Foundation of China (No. 81860826).
Author information
Authors and Affiliations
Contributions
YZQ is the principal manuscript writer, designed the study, and wrote the manuscript. XYF and LKK contributed to the data analysis and data interpretation. LLJ contributed to the revision of the manuscript. All the authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Conflict of interest
There are no competing interests to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yan, Z., Xu, Y., Li, K. et al. Association between genetically proxied lipid-lowering drug targets, lipid traits, and amyotrophic lateral sclerosis: a mendelian randomization study. Acta Neurol Belg 124, 485–494 (2024). https://doi.org/10.1007/s13760-023-02393-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13760-023-02393-w