Abstract
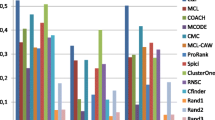
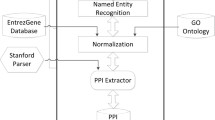
Complex biological systems are often represented as networks and studied computationally. In protein–protein interaction networks, interactions give rise to certain compounds known as protein complexes. Identifying functional protein complexes is an emerging field of study in system biology. Several machine learning methods have been proposed so far to detect functionally enriched protein complexes responsible for specific biological functions or diseases. We present an empirical study on the different unsupervised approaches towards the identification of such complexes. We report performance of seven popularly known methods against four benchmark datasets in terms of six evaluation parameters. We also highlight some issues and challenges prevailing in this field of research.







Similar content being viewed by others
References
Ali W, Deane C, Reinert G (2011) Protein interaction networks and their statistical analysis. Handbook of Statistical, Systems Biology. pp 200–234
Asur S, Ucar D, Parthasarathy S (2007) An ensemble framework for clustering protein-protein interaction networks. Bioinformatics 23(13):i29–i40
Boulton SJ, Anton G, Jérôme R, Philippe V, Nick D, Hill DE, Vidal M (2002) Combined functional genomic maps of the c. elegans dna damage response. Science 295(5552):127–131
Chen Y, Jacquemin T, Zhang S, Jiang R (2014 Prioritizing protein complexes implicated in human diseases by network optimization. BMC Syst Biol 8(S-1):S2
Fields S, Song O (1989) A novel genetic system to detect protein–protein interactions. Nature 340(6230):245–246
Gambette P, Guénoche A et al (2011) Bootstrap clustering for graph partitioning. RAIRO. Oper Res 45(4):339–352
Ganegoda GU, Wang J, Wu FX, Li M (2014) Prediction of disease genes using tissue-specified gene-gene network. BMC Syst Biol 8(Suppl 3):S3
Gavin AC, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM et al (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415(6868):141–147
Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dümpelfeld B et al (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440(7084):631–636
Grira N, Crucianu M, Boujemaa N (2004) Unsupervised and semi-supervised clustering: a brief survey. A review of machine learning techniques for processing multimedia content. In: Report of the MUSCLE European Network of Excellence (FP6)
Guanming W, Feng X, Stein L (2010) Research a human functional protein interaction network and its application to cancer data analysis. Genome Biol 11:R53
Guénoche A (2011) Consensus of partitions: a constructive approach. Adv Data Analysis Classif 5(3):215–229
Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K et al (2002) Systematic identification of protein complexes in saccharomyces cerevisiae by mass spectrometry. Nature 415(6868):180–183
Krogan NJ, Cagney G, Haiyuan Y, Zhong G, Guo X, Ignatchenko A, Li J, Shuye P, Datta N, Tikuisis AP et al (2006) Global landscape of protein complexes in the yeast saccharomyces cerevisiae. Nature 440(7084):637–643
Li M, Chen J, Wang J, Bin H, Chen G (2008a) Modifying the dpclus algorithm for identifying protein complexes based on new topological structures. BMC Bioinform 9(1):398
Li M, Wang J, Chen J (2008b) A fast agglomerate algorithm for mining functional modules in protein interaction networks. In: BioMedical Engineering and Informatics. BMEI 2008. International Conference on, vol. 1. IEEE, pp 3–7
Li X, Min W, Kwoh CK, Ng SK (2010) Computational approaches for detecting protein complexes from protein interaction networks: a survey. BMC Genomics 11(Suppl 1):S3
Lin C, Cho Y, Hwang W, Pei P, Zhang A (2007) Clustering methods in protein-protein interaction network. Knowledge Discovery in Bioinformatics: techniques, methods and application. pp 1–35
Nepusz T, Haiyuan Y, Paccanaro A (2012) Detecting overlapping protein complexes in protein-protein interaction networks. Nature Methods 9(5):471–472
Pizzuti C, Rombo SE, Marchiori E (2012) Complex detection in protein-protein interaction networks: a compact overview for researchers and practitioners. In: Evolutionary Computation, Machine Learning and Data Mining in Bioinformatics. Springer, pp 211–223
Rahman MS, Ngom A (2013) Fac-pin: Fast agglomerative clustering method for functional modules and protein complex identification in pins. In: Computational Advances in Bio and Medical Sciences (ICCABS), 2013 IEEE 3rd International Conference on. IEEE, pp 1–6
Ren J, Wang J, Li M, Wang L (2013) Identifying protein complexes based on density and modularity in protein-protein interaction network. BMC Syst Biol 7(4):1–15
Ruan J, Zhang W (2008) Identifying network communities with a high resolution. Phys Rev E 77(1):016104
Tong AHY, Evangelista M, Parsons AB, Hong X, Bader GD, Pagé N, Robinson M, Raghibizadeh S, Hogue CWV, Bussey H et al (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294(5550):2364–2368
Tong AHY, Lesage G, Bader GD, Ding H, Hong X, Xin X, Young J, Berriz GF, Brost RL, Chang M et al (2004) Global mapping of the yeast genetic interaction network. Science 303(5659):808–813
Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan Maithreyan, Pochart Pascale et al (2000) A comprehensive analysis of protein-protein interactions in saccharomyces cerevisiae. Nature 403(6770):623–627
Von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P (2002) Comparative assessment of large-scale data sets of protein–protein interactions. Nature 417:399–403
Wang J, Li M, Deng Y, Pan Y (2010) Recent advances in clustering methods for protein interaction networks. BMC Genomics 11(Suppl 3):S10
Wang J, Ren J, Li M, Wu FX (2012) Identification of hierarchical and overlapping functional modules in ppi networks
Wang X, Wang Z, Ye J (2011) Hkc: An algorithm to predict protein complexes in protein-protein interaction networks. J Biomed Biotechnol
Zhang XF, Dai DQ, Ou-Yang L, Yan H (2014) Detecting overlapping protein complexes based on a generative model with functional and topological properties. BMC Bioinformatics 15(1):186
Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T et al (2001) Global analysis of protein activities using proteome chips. Science 293(5537):2101–2105
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sharma, P., Ahmed, H.A., Roy, S. et al. Unsupervised methods for finding protein complexes from PPI networks. Netw Model Anal Health Inform Bioinforma 4, 8 (2015). https://doi.org/10.1007/s13721-015-0080-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-015-0080-7