Abstract
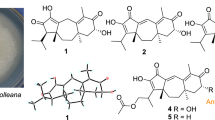
Three new heptelidic acid derivatives (1–3) including two new dimeric esters and two known heptelidic acid analogues (4 and 5) were isolated from the solid culture of mushroom Lentinellus ursinus. The structures of new compounds were confirmed by the analysis of NMR and HRESIMS spectroscopic data. The biosynthetic origin of compounds 1–5 was postulated. Compounds 1–5 exhibited no antibacterial activity against Staphylococcus aureus and Escherichia coli at the dose of 100 μM.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The mushrooms in the genus of Lentinellus are white rot, wood decay, and characterized with rough-walled and amyloid spores. Eighteen species and varieties of Lentinellus have been described all over the world. There have been eleven species reported in China [1, 2].
Mushroom-derived natural products draw much attention of chemists and biologists due to their diverse structural skeletons and interesting biological activities [3]. Mushrooms have been known as a prolific source of structurally diverse sesquiterpenes [4,5,6,7]. Heptelidic acid and its analogues are a group of significant secondary metabolites with interesting biological activities, such as cytotoxic, antimicrobial, antimalarial activities [8,9,10]. So far, heptelidic acid and its derivatives have been reported from the genus of fungi Lentinellus, Gliocladium, Chaetomium, Trichoderma, Xylaria, Phyllosticta, and Acremonium [8, 9, 11,12,13]. In our ongoing search for new sesquiterpenes from mushrooms, the EtOAc extract from the solid culture of Lentinellus ursinus was investigated. To date, only one sesquiterpene lentinellic acid was reported from the liquid culture of L. ursinus [14]. In this study, five heptelidic acid derivatives including a new acetylated compound and two new dimeric sesquiterpenoid esters were isolated from the fungus L. ursinus. Herein, we report the isolation, structure elucidation, and antibacterial activity evaluation of new compounds 1–3 (Fig. 1).
2 Results and Discussion
The fungus L. ursinus was fermented on rice medium. The EtOAc extract of its rice culture was subjected to silica gel, ODS, Sephadex LH-20, and HPLC chromatography to afford three new compounds 3-O-acetylheptelidic acid A (1) lentisinic acid A (2) and B (3), and two known compounds hydroheptelidic acid (4) [12] and xylaric acid D (5) [11].
3-O-Acetylheptelidic acid A (1) was obtained as a colorless oil. Its molecular formula was determined to be C17H24O7 by the HRESIMS (m/z 363.1414 [M+Na]+). The 1H, 13C NMR, and HSQC data (Table 1) displayed a singlet methyl (δC/δH 20.9/2.00), two doublet methyls [δC/δH 15.9/0.75 (d, J = 6.9 Hz), 21.9/0.97 (d, J = 6.9 Hz)], four methylenes containing two oxygenated methylenes [δC/δH 59.2/4.82 (d, J = 12.1 Hz), 4.59 (d, J = 12.1 Hz); 76.2/4.45 (d, J = 9.9 Hz), 4.06 (d, J = 9.9 Hz)], four methines, a trisubstituted double bond (δC/δH 148.8/6.80 (d, J = 9.9 Hz), 130.8), an oxygenated quaternary carbon (δC 76.3), and three carbonyl carbons (δC 169.2, 172.5, 178.8). These data indicated that 1 shares the same sesquiterpenoid skeleton with hydroheptelidic acid (4) [12], except for an additional acetyl group. The 1H–1H COSY correlations of H-11/H-12, H-5/H-11/H-10/H2-9/H2-8, H-10/H-13/H3-14(15), and key HMBC correlations of H2-3 with C-1 and C-2, H-12 with C-1, C-2, C-3, and C-5, H-5 with C-6 and C-4, H2-7 with C-4, C-6, and C-5, H-8 with C-5, C-6 and C-7, H2-3 (δH 4.82, 4.59) and the methyl group (δH 2.00) with the carbonyl carbon (δC 172.5) indicated the attachment of the acetyl group at C-3 definitely assigned the structure of 1 (Fig. 2). The strong NOE correlations of H-11 with H2-3 supported the E-configuration for the C(2)=C(12) bond. The NOE correlations of H-11 with H-13, H3-14 and H3-15, H-5 with H-10, together with the larger coupling constants (3J5, 11 = 11.0 Hz) between H-5 and H-11 attributed the a-orientation for H-11 and β-orientation for H-5 and H-10 (Fig. 3). The obvious NOE correlation of HO-6 (δH 5.38 in DMSO-d6) with H-6 (δH 2.06 in DMSO-d6) assigned the β configuration of HO-6. Considering the same biosynthetic pathway for the heptelidic acid derivatives, the absolute configuration of 1 was deduced to be 5S, 6S, 10R, 11S.
The molecular formula of 2 was determined to be C30H42O11 with 10 degrees of unsaturation, as determined by HRESIMS (m/z [M+Na]+ 601.2615). The 1H, 13C NMR, and HSQC data of 2 revealed the presence of four doublet methyls [δC/δH 15.6/0.94 (d, J = 6.9 Hz), 15.9/0.79 (d, J = 6.9 Hz), 21.6/1.00 (d, J = 6.9 Hz), 21.9/0.98 (d, J = 6.9 Hz)], eight methylenes containing four oxygenated methylenes [δC/δH 57.2/4.30 (d, J = 12.4 Hz), 4.16 (d, J = 12.4 Hz); 62.7/5.23 (d, J = 14.6 Hz), 5.06 (d, J = 14.6 Hz); 66.3/5.18 (d, J = 12.3 Hz), 4.59 (d, J = 12.3 Hz); 76.3/4.48 (d, J = 9.9 Hz), 4.08 (d, J = 9.9 Hz)], eight methines, two pairs of trisubstituted olefinic carbons [δC/δH 147.0/7.28 (d, J = 4.6 Hz), 130.9; 146.1/6.71 (d, J = 10.7 Hz), 134.7], two oxygenated quaternary carbon (δC 74.0, 76.4), and four carbonyl carbons (δC 168.4, 174.4, 168.5, 179.5) (Table 2). The HMBC and 1H–1H COSY spectral analysis (Fig. 2) confirmed the presence of two sesquiterpenes moieties, respectively, corresponding to heptelidic acid [15] and hydroheptelidic acid (4) [12]. The heptelidic acid residue was determined by 1H–1H COSY correlations of H-11/H-12, H-5/H-11/H-10/H2-9/H2-8, H-10/H-13/H3-14(15), and key HMBC correlations from H2-3 to C-1 and C-4, H-5 to C-4 and C-6, H2-7 to C-5, C-6 and C-8, H-12 to C-1, C-2, C-3, C-5 and C-10. The remaining signals were belonging to the hydroheptelidic acid moiety was assigned by 2D NMR spectral data. Finally, an ester bond was assigned between C-7 and C-1′ to satisfy the requirement of the molecular weight, which was also supported by the HMBC correlations of H-7 (δH 5.18, 4.59) with C-1′ (δC 168.4). NOE correlations of H-5/H-10, H-11/H2-7, H-11/H-13, H-3′/H-11′, H-11′/H3-14′, H-5′/H-10′, together with the lager coupling constants of C-5 and C-11 (3J5,11 = 12.6 Hz), H-5′ and H-11′ (3J5′,11′ = 10.5 Hz) confirmed the relative configuration in 2 as described (Fig. 4).
Lentisinic acid B (3) has the molecular formula C30H40O11 with 11 unsaturation degrees, as determined from HRESIMS at m/z [M+Na]+ 583.2517. The 1H and 13C NMR data of 3 revealed a similar dimeric structural feature as that of 2, except for the presence of two extra olefinic carbons (δC 125.3, 164.7) in 3, and the absence of a methine and an oxygenated quaternary carbon in 2. Two olefinic carbons were assigned at C-5′ and C-6′ by key HMBC corrections of H-12′ with C-5′, H2-8′ with C-5′ and C-6′, H2-7′ with C-5′ and C-6′. Further analysis of 2D NMR data (Figs. 2, 4) confirmed the structure of 3.
All isolated compounds were tested for antibacterial activities against Staphylococcus aureus and Escherichia coli. None of them showed antibacterial activity at 100 μM.
The sesquiterpene lactone of compounds 1–5 are structurally related to each other and might be originated from heptelidic acid. A proposed biogenetic pathway for these compounds is shown in Fig. 5. Heptelidic acid derived from the 1,10- and 1,6-cyclization of FPP [15] was converted into 6 by hydrolysis and oxidation cleavage. 4 was formed from 6 by dehydration, and further transformed into 1 and 5 by acetylation or dehydration, respectively. The intermidate 7 derived from heptelidic acid was further reacted with 4 and 5 to give new dimeric compounds 2 and 3, respectively.
In conclusion, three previously undescribed cadinane-type sesquiterpenes including one acetylated heptelidic acid derivative (1), two dimeric esters (2–3), and two known heptelidic acid analogues (4–5), were isolated from the solid culture of L. ursinus. The current study enriches the secondary metabolites from this mushroom.
3 Experimental
3.1 General Experimental Procedures
HPLC separation was conducted on Agilent 1200 HPLC system equipped with Agilent G1315D DAD detector, using a YMC-Pack ODS-A column (5 μm; 9.4 × 250 mm). NMR spectra were measured on a Bruker Avance-500 spectrometer using solvent signals (CD3OD, δC/δH 49.00/3.31; CDCl3, δC/δH 77.16/7.26, DMSO-d6, δC/δH 39.52/2.50) as references. Mass spectra were obtained on an Agilent Accurate-Mass-Q-TOF LC/MS 6520 spectrometer. Optical rotations were recorded on a polarimeter with sodium light (589 nm) by using a Perkin-Elmer 241 polarimeter. UV and IR spectra were recorded on a Thermo Genesys-10S UV–Vis spectrophotometer and a Nicolet IS5FT-IR spectrophotometer, respectively.
3.2 Fungal Material
The strain of L. ursinus was isolated from the fruiting bodies of mushroom L. ursinus collected in Meilixueshan (Yunnan, China) by Junjie Han, and identified on the basis of the morphological characteristics and ITS sequences. The strain was cultured on PDA plates at 25 °C for 14 days. Agar plugs were inoculated into a 250 mL Erlenmeyer flask containing 100 mL PDB medium cultured at 25 °C on a rotary shaker at 180 rpm for 14 days. Large-scale cultivation was carried out in 20 × 500 mL Fernbach culture flasks each containing 80 g of rice and 100 mL of distilled water. Each flask was inoculated with 5 mL of culture medium and incubated at 25 °C for 40 days in dark.
3.3 Extraction and Isolation
The cultivated rice substrate was extracted repeatedly with EtOAc (3 × 10 L) at room temperature and 20.2 g crude extract was obtained by evaporating solvent under vacuum. The crude extract was subjected to silica gel column chromatography (CC) using CH2Cl2–MeOH (100:1, 80:1, 60:1, 40:1, 25:1, 10:1, 5:1, 0:1) to give fifteen fractions (Fr.1–Fr.15).
The fraction Fr.7 (4.0 g) eluted with 40:1 CH2Cl2–MeOH was separated by ODS CC eluting with MeOH–H2O gradient elution to give nineteen subfractions (Fr.7.1–Fr.7.19). The fraction Fr.7.4 (1.0 g) was purified by Sephadex LH20 CC eluted with 80% MeOH–H2O, followed by PR-HPLC (20% MeCN in H2O) to give 4 (10.0 mg, tR = 37.8 min). The fraction Fr.7.6 (200.0 mg) was purified by PR-HPLC (28% MeCN in H2O) to afford 1 (10.8 mg, tR = 26.2 min) and 5 (13.3 mg, tR = 30.0 min). Compound 2 (3.5 mg, tR = 18.5 min) and 3 (6.2 mg, tR = 22.4 min) were purified by PR-HPLC from Fr.7.11 (45% MeCN in H2O) and Fr.7.13 (50% MeCN in H2O), respectively.
3.4 Spectroscopic Data
3.4.1 3-O-Acetylheptelidic acid A (1)
Colorless oil; [α] 25D 62.9 (c 0.1 MeOH); UV (MeOH) λmax (log ε) 222 (3.3) nm; IR(neat) vmax 3432, 2959, 1778, 1719, 1465, 1310, 1233, 1170, 1019 cm−1; positive HRESIMS m/z [M+Na]+ 363.1412 (calcd for C17H24O7Na, 363.1414).
3.4.2 Lentisinic acid A (2)
Light yellow powder; \([\upalpha ]_{{\text{D}}}^{{25}}\) 23.0 (c 0.1 MeOH); UV (MeOH) λmax (log ε) 217 (3.4) nm; IR(neat) vmax 3447, 2958, 1734, 1717, 1472, 1388, 1238, 1170, 1018 cm−1; positive HRESIMS m/z [M+Na]+ 601.2615 (calcd for C30H42O11Na, 601.2619).
3.4.3 Lentisinic acid B (3)
Light yellow powder; \([\upalpha ]_{{\text{D}}}^{{25}}\) 71.5 (c 0.2 MeOH); UV (MeOH) λmax (log ε) 222 (3.4) nm; IR(neat) vmax 3432, 2959, 1778, 1719, 1465, 1370, 1233, 1170, 1019 cm−1; positive HRESIMS m/z [M+Na]+ 583.2517 (calcd for C30H40O11Na, 583.2514).
3.5 Antimicrobial Assay
The antimicrobial assay was conducted as our previous described method [16]. The bacterial strains S. aureus (ATCC 6538) and E. coli (ATCC 25922) were grown in Lysogeny Broth (LB) medium. The inhibition rate was calculated and plotted versus test concentrations to afford the MIC. MIC values were defined as the minimum concentration of compound that inhibited visible microbial growth.
References
P.M. Kirk, P.F. Cannon, D.W. Minter, J.A. Stalpers, Ainsworth and Bisby’s Dictionary of the Fungi, 10th edn. (CAB International, Wallingford, 2008), pp. 1–771
T.H. Li, W.Q. Deng, C.W. Deng, B. Song, J. Fungal. Res. 10, 130–132 (2012)
D.K. Rahi, D.J. Malik, Mycol. 2016, 1–18 (2016)
S.J. Wang, L. Bao, F. Zhao, Q.X. Wang, S.J. Li, J.W. Ren, L. Li, H.A. Wen, L.D. Guo, H.W. Liu, J. Agric. Food Chem. 61, 5122–5129 (2013)
R.H. Yin, Z.Z. Zhao, X. Ji, Z.J. Dong, Z.H. Li, T. Feng, J.K. Liu, Nat. Prod. Bioprosp. 5, 17–22 (2015)
M. Isaka, S. Palasarn, M. Sappan, S. Supothina, T. Boonpratuang, Nat. Prod. Bioprosp. 6, 257–260 (2016)
Y.Q. Wang, L. Bao, X.L. Yang, L. Li, S.F. Li, H. Gao, X.S. Yao, H.A. Wen, H.W. Liu, Food Chem. 132, 1346–1353 (2012)
Y. Yamaguchi, D. Manita, T. Takeuchi, K. Kuramochi, I. Kuriyama, F. Sugawara, H. Yoshida, Y. Mizushina, Biosci. Biotechnol. Biochem. 74, 793–801 (2010)
Y. Itoh, K. Kodama, K. Furuya, S. Takahashi, T. Haneishi, Y. Takiguchi, M. Arai, J. Antibiot. 33, 468–473 (1980)
Y. Tanaka, K. Shiomi, K. Kamei, M. Sugoh-Hagino, Y. Enomoto, F. Fang, Y. Yamaguchi, R. Masuma, C.G. Zhang, X.W. Zhang, S. Omura, J. Antibiot. 51, 153–160 (1998)
S. Yan, S.Y. Li, W. Wu, F. Zhao, L. Bao, R. Ding, H. Gao, H.A. Wen, F.H. Song, H.W. Liu, Chem. Biodivers. 8, 1689–1700 (2011)
L.A. Calhoun, J.A. Findlay, J.D. Miller, N.J. Whitney, Mycol. Res. 96, 281–286 (1992)
J. Kawashima, F. Ito, T. Kato, M. Niwano, H. Koshino, M. Uramoto, J. Antibiot. 47, 1562–1563 (1994)
A. Stärk, T. Anke, U. Mocek, W. Steglich, A. Kirfel, G. Will, Z. Naturforsch. C. 43, 177–183 (1988)
R.D. Stipanovic, C.R. Howell, Tetrahedron 39, 1103–1107 (1983)
Q.X. Wang, L. Bao, X.L. Yang, H. Guo, R.N. Yang, B. Ren, L.X. Zhang, H.Q. Dai, L.D. Guo, H.W. Liu, Fitoterapia 83, 209–214 (2012)
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (21472233 and 81673334).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Liu, L., Han, JJ., Xu, TS. et al. Three New Heptelidic Acid Derivatives from the Culture of Mushroom Lentinellus ursinus. Nat. Prod. Bioprospect. 8, 355–360 (2018). https://doi.org/10.1007/s13659-018-0168-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13659-018-0168-8