Abstract
The forage legume species biserrula (Biserrula pelecinus) is among a few forage species with low enteric methane emission when fermented by rumen microbes and it is unclear whether metabolites in biserrula play a role in this. The hypothesis of this study was that specific metabolites in biserrula are candidate biomarkers for and associated with low methanogenesis. We characterized the metabolomic profiles of 30 accessions of biserrula or its core collection using nuclear magnetic resonance and liquid chromatography mass spectrometry. These profiles were then analyzed for association with methanogenesis potential in the rumen. Metabolomic profile was predictive of methanogenesis. Forty-seven putative metabolites were identified by liquid chromatography mass spectrometry that were highly expressed (P < 0.05) in the associations with low methanogenic potential. Fragmentation analysis of a subset of these metabolites suggested saponins, with one feature tentatively identified as an ursolic or oleanolic terpene glucoside, consistent with the nuclear magnetic resonance data. Accessions with Eritrean geographic origin were metabolomic outliers. Here, we show for the first time that some metabolites contribute to the methane mitigation effects of biserrula. If in vivo results confirm this, the environmental impact of this study would be the availability of biomolecules for livestock vaccination to mitigate methane emission resulting in an economic impact of lower cost of production in countries with a price on environmental emissions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
CH4 is a common byproduct of anaerobic microbial fermentation of forage in the rumen (Fig. 1a). Ruminants are vital to food production in most countries of the world, and in Australia and New Zealand, their products are important contributors to economic and social wellbeing. Enteric CH4 emission occurs as a result of microbial fermentation of feed components (Johnson and Johnson 1995). CH4 produced by domesticated ruminants represents a loss of 2–15% of the gross energy intake (Holter and Young 1992), thus being an important inefficiency in ruminant production systems (Moss et al. 2000). To abate CH4 emission and increase the efficiency of digestion in ruminants, the focus is to divert some of the energy lost as CH4 into animal products (Mitsumori and Sun 2008). Enteric fermentation can vary widely depending on factors such as type of the animal, feed intake, the type and quality of feed, environment, adding dietary fat, feed additives, rumen microbiota, and, indirectly, body weight of the animal through feed intake (Moss et al. 2000).
There are urgent needs to understand the various factors affecting variability in enteric CH4 production to decrease the uncertainty in GHG emission inventories and to identify viable GHG reduction strategies (Sejian et al. 2011). Improving pasture quality is viewed as a means of reducing CH4 emissions because animal productivity may be enhanced (i.e., lowering CH4 emissions per unit of animal product) with less dietary energy lost as CH4 due to reduced fiber content. In New Zealand, Friesian and Jersey dairy cows grazing sulla (Hedysarum coronarium), a condensed tannins-containing legume, emitted less CH4 per unit of dry matter than cows grazing perennial ryegrass pasture (Woodward et al. 2002). Further, the antimicrobial properties of essential oils have been shown through in vitro and in vivo studies to inhibit a number of bacteria and yeasts and to control fermentation gases, livestock waste odors, and human pathogenic bacteria (Helander et al. 1998; Lee et al. 2002; Wallace et al. 2002). Experiments conducted in vitro (Ulyatt et al. 2002) and in vivo (Bayaru et al. 2001) show that addition of organic acids to the diet reduces CH4 production.
In a recent study, a broad range of dietary pasture and forage species and their effect on CH4 production in ruminants was reported (Banik et al. 2013). The most promising among these species was biserrula (Biserrula pelecinus), an important annual legume for the wheat belt of southern Australia (Fig. 1b).
Recently, a core collection of 30 accessions of biserrula has been developed from the genetic resource of 279 accessions in the Australian Trifolium Genetic Resource Centre (Perth, Western Australia), held by the Department of Agriculture and Food Western Australia (DAFWA) (Ghamkhar et al. 2012). Preliminary LCMS analysis of a subset (12) of the biserrula accessions has previously been reported by Ghamkhar et al. (2016). Here, we report the use of metabolomics techniques to investigate the effect of all accessions of the core collection of biserrula on CH4 production when supplied to a rumen fluid suspension in vitro. In the past, some phytochemicals have been suggested to have a direct toxic effect on methanogens (e.g., condensed tannins) or protozoa (e.g., saponins) (Patra and Saxena 2009) and the heterogeneous group of compounds known as essential oils also show correlation with methanogenesis (Martin et al. 2010). Recent studies suggested plant secondary compounds as a cue in methanogenic potential of biserrula (Swinny et al. 2015; Banik et al. 2016). Identification of metabolites in this study will provide the same, similar, or different candidate phytochemical targets for development of novel classes of methanogen inhibitors. These measurements and correlations should assist plant breeders to make better choices in plant selection, in particular targeting CH4 mitigation from livestock.
The aims of the current study were to:
-
1.
Compare the metabolomics profiles of the accessions of the biserrula core collection and their association with CH4 emission using an in vitro fermentation technique (IVFT); and
-
2.
Identify candidate metabolites that can reduce methane production in the rumen and suggest future environmentally and phytochemically targeted plant breeding options in biserrula.
2 Materials and methods
2.1 Plant material
The core collection of biserrula (Biserrula pelecinus L.), made up of 30 accessions conserved in the ATGRC, and two controls, red clover (Trifolium pratense L. cv. Redquin) and subterranean clover (Trifolium subterraneum L. cv. Woogenellup) (Table 1) were sampled and screened for their in vitro CH4 output analysis as described by (Banik et al. 2013). Briefly, 47 plants of 30 accessions of the biserrula core collection with half of them replicated, and two control species, subterranean and red clover, were grown in pots in a glasshouse, and plant material was collected for in vitro analysis of methanogenic potential at 77 days post-sowing. The herbage consisting of leaf and stem 3 cm above ground level when elevated were harvested from individual plants, and the material was freeze-dried in a bench-top freeze dryer (BenchTop, VirTis/SP Scientific, Warminster, PA) and ground in a grinding mill (Glen Creston, Stanmore, UK) to pass through a 1.0-mm screen. Material was stored at room temperature in sealed containers until analysis by Banik et al. (2013).
The same ground plant material was used for metabolomics analysis except that accession numbers 11, 21, 29, and 37, the latter two being replications of the same accession were missing and not available for this analysis (Table 1). However, 27 accessions of biserrula from different geographic origins and a diverse range of CH4 output as well as the two clover controls were still available for metabolomics analysis as shown in Table 1.
2.2 In vitro fermentation technique
Data from Banik et al. (2013) were sorted and processed for use as the base information for the 27 accessions of biserrula and the control species (Table 1). Briefly, plants were harvested at 10 weeks. The edible parts of the plants was freeze-dried in a bench-top freeze dryer (BenchTop, VirTis/SP Scientific, Warminster, PA) and ground in a grinding mill (Glen Creston, Stanmore, UK). Fermentability was examined in an in vitro batch fermentation system commonly used to examine plant fermentability (Bodas et al. 2008). One day before the testing, 0.1 g of plant material was weighed and transferred to an anaerobic chamber (Coy Laboratory Products Inc., Grass Lake, MI) maintained at 39 °C and supplied with 80% N2: 10% CO2: 10% H2 to expel oxygen from the tubes. Before the commencement of fermentation, rumen fluid was collected from two fistulated sheep fed a general maintenance diet consisting of lupins and oaten chaff 2 h after feeding.
After straining, rumen fluid was pooled, transferred into the anaerobic chamber, and buffered to pH 7.2. Then 10 mL of this mix was dispensed into prepared tubes. Inside the chamber, the tubes were sealed and incubated for 24 h at 39 °C, with constant shaking at 50 rpm. Tubes were then placed in a water bath at 39 °C. After measuring gas pressure, 5 mL of headspace gas was used for subsequent methane analysis by gas chromatography (GC). Methane concentration in the gas sample was determined using a Micro-GC with a PoraPLOT Q 0.25 mm × 10 m column (Varian/Agilent Technologies, Australia Pty Ltd., Mulgrave, Vic.), at 50 °C, a thermal conductivity detector, and Galaxie software (Varian, Australia). The methanogenic potential of the plant was expressed as total methane produced (mL/g DM incubated, (Patra et al. 2006)).
2.3 Mass spectrometry
LCMS was based on the methodology described by the authors (Ghamkhar et al. 2016), but in this experiment, a much larger sample of 30 accessions was examined with the following steps:
2.3.1 Extraction
In brief, ground, freeze-dried samples (100 mg) were extracted in 1 mL 80% methanol:water, by sonication for 10 min. After centrifugation, 200 μL of supernatant from each sample was placed into an HPLC vial for LCMS analysis. This LCMS sample was stored at 4 °C overnight prior to analysis. The extraction process was repeated on the pellet for a second time. Combined extracts were then reduced in volume to less than 300 μL by evaporation under N2 gas whilst heating at 30 °C. Samples were frozen at − 80 °C and then freeze-dried using a Christ Freeze Drying System (ice condenser, − 55 °C; vacuum: 0.030 mbar) (Martin Christ Gefriertrocknungsanlagen GmbH, Germany). Freeze-dried samples were stored at − 80 °C over desiccant. Duplicates were prepared for three samples to test method reproducibility.
2.3.2 LCMS sample analysis
LCMS data was acquired in negative ionization mode at high resolution. Negative Fourier transform mode lenses were tuned to m/z of 739.20 by infusion of sample number 3 when teed with mobile phase at initial chromatographic conditions.
HPLC separation was achieved using a 150 mm × 2.1 mm ThermoFisher 1.9 μm C18 HPLC column (Thermo Fisher Scientific, Australia) fitted to an Agilent series 1290 high-performance liquid chromatograph (binary pump, cooled auto-sampler maintained at 4 °C, column heater maintained at 30 °C and photodiode array detector) utilizing a gradient elution, (98 to 0%A over 20 min followed by a 5 minute wash with 100% B before returning to the initial gradient conditions for 4 minutes; A: 0.1% formic acid in water; B: 0.1% formic acid in acetonitrile, at 0.3 ml min−1). The HPLC was connected to a ThermoFisher LTQ Velos orbitrap mass spectrometer (Thermo Fisher Scientific, USA), operating with a heated electrospray ionization probe operating in a negative FT mode over 100–2000 m/z. The heated capillary was maintained at 300 °C with a source heater temperature of 350 °C and the sheath, auxiliary, and sweep gases at 37, 13, and 7 units respectively. The source voltage was 3.3 kV.
MS2 and MS3 analyses were carried out on selected samples using a data-dependent approach with accurate mass in MS1 and MS2 set at resolution of 15,000 for MS2 and MS3. Dynamic exclusion was engaged with a repeat duration of 30 s and an exclusion duration of 180 s.
2.3.3 LCMS data analysis
Initial data processing was carried using Expressionist Refiner MS (Genedata, Basel, Switzerland) to align MS data, carry out noise reduction, and for peak picking. Cluster analysis was undertaken to group isotope peaks and singletons removed. The cluster volume was analyzed statistically in Expressionist Analyst (Genedata, Basel, Switzerland). Samples were classified into groups according to methanogenic potential and by country of origin (Table 1). Principal components analysis (PCA) and hierarchical cluster analysis (HCA)—Manhattan—were used to explore the relationship between the accessions. For both methods valid values were set to a minimum of 20%. To investigate specific features that were elevated in the low methanogenic potential group compared to the high group, a t test was used with 30% valid values per group with bootstrapping (10 repeats). P values were adjusted using Bonferonni’s correction (P = 0.05 equivalent to P = 7.7e-6). In order to identify specific metabolomic features that could be contributing to the low methanogenic potential of the biserrula accessions, the LCMS data was re-examined. The clover samples were omitted from analysis so that only the high and low biserrula accessions were considered. These were then analyzed by a t test. The features that were highly statistically significant and that had an effect size of ten or greater (elevated in the low CH4 accessions) were selected as the most likely candidate features with CH4 lowering potential.
2.4 NMR sample preparation
Freeze-dried sample extracts were re-dissolved for NMR analysis in 600 uL of DMSO-d6 by vortexing and sonicating. Samples were centrifuged for 10 min at 14,000 rpm and 20 °C. A total of 550 uL was transferred to NMR tubes for analysis.
2.4.1 NMR analysis
NMR data was acquired on a Bruker 700-MHz instrument equipped with a cryoprobe and an autosampler. The noesypr1d pulse sequence was used with a spectral width of 18.6 ppm, 256 scans (8 dummy scans), and a total acquisition time of 1.26 s. The data were processed in Topspin 3.2 (Bruker Biospin, Germany) with a line broadening of 0.3 Hz. Spectra were referenced to DMSO (2.5 ppm). 2D NMR data was acquired for samples 4 (high) and 14 (low). The pulse sequences used were socygpqf, hsqcedetgpsisp 2.2, and hmbcgplpndqf.
2.4.2 NMR data analysis
Proton spectra were imported into MatLab (R2015b, Mathworks) using a modified Prometab script, as previously described (Rochfort et al. 2015). Spectra were baseline corrected and normalized (total spectral area set to one) in PLSToolbox (Ver 8.0.2, Eigenvector Research). The spectra were analyzed by PCA with mean centering pre-processing. This analysis confirmed that the replicate samples (for 17, 36, and 46) clustered closely to each other and revealed the spectrum of sample 2 to be an outlier due to poor spectral quality. The spectra from replicates and sample 2 were removed and the residual solvent (2.48–2.55 ppm) was deleted. This data was used in all subsequent PCA analyses. Samples were classified into groups according to methanogenic potential (Table 1).
3 Results and discussion
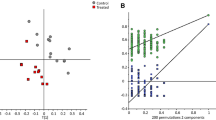
Initially, the “high” and “low” categories in methanogenic potential assigned in Table 1 involved the drawing of a somewhat arbitrary line through a continuous distribution. However, analysis of the LCMS data by unsupervised modeling techniques including PCA and HCA revealed clear trends in the data. In the PCA scores plot (data not shown), each accession was classified as having low or high (within biserrula’s already low range of methanogenesis), and very high (in two clovers) methanogenic potential based on the in vitro CH4 output (Banik et al. 2013). In general, the accessions cluster based on methanogenic potential with low and high groups separated on PC1 and the very high group separated on PC2. The clear outlier here was sample 3, which did not appear to cluster according to CH4 group. Generally, the HCA showed that the clustering of the accessions by their methanogenic potential was in line with their origin (Fig. 2).
Hierarchical clustering of liquid chromatography mass spectrometry (LCMS) data in the accessions of biserrula and the two clover control species. Accessions are clustered based on their color-coded methane output (arrow indicating accession 3). Countries of origin of the accessions are projected on the clusters
Initial analysis of NMR data with all accessions and control clover species using PCA revealed a similar pattern to the LCMS results with low and high accessions separating on PC1 and very high accessions separating on PC2 with the expected graduation across the two groups (not shown here). Accession 3 still stayed with its contrasting CH4 output group.
Results obtained from both LCMS and NMR PCA suggest that high and low CH4 output accessions are, to a large extent, two chemically distinct groups within the core collection of biserrula. Further, hierarchical cluster analysis indicates that within the high CH4 output group of accessions, the metabolic profile of accessions from Eritrea is clearly distinct from accessions of the northern Mediterranean region. This supports the previous suggestion (Ghamkhar et al. 2012) that the Eritrean accessions are a differentiated group of accessions within B. pelecinus germplasm and distinctly different from the other accessions of biserrula, in agro-morphological traits, eco-geographical distribution, and genomic profile. However, this study suggests that they still share some metabolites with some accessions from Morocco and Canary Islands, regions geographically isolated from the northern Mediterranean region.
It can be seen that the outlier in PCA (accession 3) clusters with its respective eco-geographical origins after the HCA (Fig. 2), while the latter analysis identified 6484 features in metabolomics data. It separated the control species more clearly from the biserrula accessions (Fig. 2). It also separated the accessions with higher CH4 output from the ones with lower CH4 output, although there were three exceptions. Accession number 20 (same accession as 41) with a high CH4 output was clustered with the lower CH4 output cluster. Also, as in the PCA, accession 3 was grouped with the high emitters but it was clustered with its geographical group (Fig. 2). In general, this analysis, although on metabolites, indicates a pattern from high CH4 output to low CH4 output accessions. When the geographical site of the accessions is added to the equation, Eritrea is identified as the hotspot of accessions with high CH4 output. A change in CH4 output, from high to low, was observed in the collection sites from Africa to Europe via Morocco and the Canary Islands. The dendrogram was sub divided into four separate clusters (Fig. 2). Clovers were clearly separate from all biserrula accessions (cluster “A” in Fig. 2). All accessions from Eritrea clustered, firstly, together and then, distinctly with some Moroccan and Canary Islands accessions. The rest of the accessions could be geographically categorized to two regions: the western Mediterranean region or cluster C (Morocco, Canary Islands, Spain, France, and Italy), and the eastern Mediterranean region or cluster D (Greece and Israel) (Fig. 2).
Among the low methanogenic accessions, Greek accessions have a similar metabolic profile with the Israeli accessions as they form a cluster together. Using agro-morphological characteristics and molecular diversity profiles, Ghamkhar et al. (2012) concluded that accessions from the western and eastern Mediterranean regions can be distinguished as two separate categories. In this study, the two regions are distinct in their metabolomics profile as well, although Eritrean accessions are, again, an exception. However, within cluster B, the East-West separation is also true.
The discovery of 47 candidate metabolites (Table 2) that may affect CH4 output in the rumen is a starting point for further investigation into these candidate metabolites as markers for this trait in other pasture species. It also provides an opportunity to discover candidate genes for controlling this trait in a reverse genetics approach, from metabolites to the transcriptome and eventually the genome by investigating enzymes that control the biochemical pathways where these markers are most active/present. Ursolic acid, a candidate marker of interest in this study, is a pentacyclic triterpene saponin and a phytosterol. It is a biochemical component with cytotoxic activity and also used as an emulsifier in pharmaceuticals, cosmetics, and foods, similar to pomolic acid found in the wax coats of apples (He and Liu 2007). Both ursolic acid and its isomer, oleanolic terpene, identified as the two main candidate markers for CH4 output mitigation in rumen, originate from the oleanyl cation in biosynthesis.
The loadings plot for PC1 (Fig. 3a) reveal some clear metabolic signatures that are responsible for the discrimination on PC1 and so between the low and high groups. The resonances between 0 and 2.5 ppm are typical of terpenes with methyl singlets resonating between 0.7 and 1.2 ppm. Methylene and methine resonances between 0.2 and 2.4 are separated and well resolved, typical of protons on more complex cyclic structures such as diterpenes and triterpenes. These terpene resonances are elevated in the spectra from the low CH4 accessions. The majority of the carbohydrate residues are resonating between 2.8 and 4.2 ppm with anomeric methines resonating between 3.8 and 5.5 ppm. This is a crowded region of the spectrum and there are resonances contributing to both the positive and negative loadings. The olefinic methines between 5.2 and 5.5 ppm seem to be contributing to the separation on the positive loadings. The aromatic methine resonances are elevated in the high accessions although, again, this is a crowded spectral region and there are some resonances contributing to the positive loadings as well.
a Principal component analysis loadings plot of NMR data for biserrula. The resonances on the positive side of the Y axis are elevated in the accessions with low methanogenic potential (upward arrow) and on the negative side of the Y axis in accessions with high methanogenic potential (downward arrow). Several structural features can be identified that may play a role in diversity of the methanogenic potential of the accessions: a–e. b MSn analysis of m/z 649.3957 where the parent ion (m/z 649.3957) generates 5 fragments in MS2 and the MS2 base peak (m/z 443.3534) generates m/z 425.3424 in MS3
Oleanolic acid and ursolic acid and their derivatives have anti-inflammatory and antioxidant effects as well (Pollier and Goossens 2012). In plants, specifically, they exist in the epicuticular waxes as a first defense barrier against pathogens (Heinzen et al. 1996) and may function as defense compounds against pathogens or as allelopathic agents (Szakiel et al. 2005). This suggests a potentially crucial role for these components as key biomolecular components to be added to future vaccines against high methanogenesis in the rumen, possibly through their antibacterial (Pollier and Goossens 2012) or anti-proliferative (Neto 2007) activity. However, Banik et al. (2016) suggested that rumen microflora can change or adapt over time in response to biserrula substrate; therefore, the short-term bioassay data has to be used with some caution. Ultimately, in vivo and in-field testing is required to confirm these properties.
3.1 Data mining for specific metabolites for CH4 output
A total of 47 features were identified (Table 2) that were significantly elevated in the low group (P < 0.05 and 10 fold or greater change in cluster volume). These 47 features correspond to metabolites of relatively high molecular weight, between 650 and 1310 amu. Accurate mass measurement was used to predict the most probable molecular formulae. However, even with a mass error of 3–5 ppm, allowed there are over 30 possible formulae for many of the ions. For this reason, and considering both the NMR and MS2 data, formulae were restricted to CHO composition. This allowed putative molecular formulae to be proposed.
Joblin (1999) stated that the management of H2 production in the rumen is the most important factor to be considered when developing strategies to control ruminant CH4 emissions. It should therefore be possible to reduce CH4 production by inhibiting H2-liberating reactions or by promoting alternative H2-using reactions or routes for disposing of H2 during fermentation. An achievement in reducing CH4 production in livestock farms would be the development of a vaccine containing an antigen derived from methanogenic bacteria (Gworgwor et al. 2006) and an immunogenic preparation which reduces the activity of rumen protozoa (Baker et al. 2004). Such vaccines have the potential to provide a cost-effective treatment to reduce CH4 emission and enhance animal production. The candidate metabolites are also potential precursors for future vaccine development if their inhibiting properties on methanogenic bacteria are proved.
MSn data was acquired in a data independent mode in separate analyses. Useful MSn data was acquired on only a few of the 47 potential biomarkers. The best data was obtained for m/z 649.3957. The parent ion (m/z 649.3957) generates five main fragments in MS2 (m/z 631.3890, 605.4063, 487.3436, 443.3534, and 425.3424). In addition, the MS2 base peak (m/z 443.3534) generates m/z 425.3424 in MS3. Accurate mass measurements on the fragment ions greatly facilitated the assignment of the molecular formulae of this molecule and allowed determination of the molecular formulae of the fragments. The mass and fragmentation pattern are consistent with an ursolic or oleanolic terpene glycoside. The fragmentation of this molecule follows the same general trend reported by Xia et al. (2015). In particular, the observation of the loss of CO2 to form the ion at m/z 605.4063 confirmed that the glycosylation cannot occur at this position and also confirms the presence of the carboxylic acid moiety in the molecule. The regiochemistry of the molecule cannot be defined by the MS data, hence the structure may be an ursolic or oleanolic terpene. The NMR data is consistent with this interpretation in that there are resonances in the regions appropriate for both the carboxylic acids and terpene glycosides (Fig. 3b). Indeed, the NMR data suggests there are many such molecules in the accessions. However, the NMR data is complex and no single molecule could be positively identified.
Although the remaining biomarkers remain unidentified, it seems very likely that they are also terpene glycosides. Further work will need to be done with both targeted MSn approaches and isolation of metabolites for NMR structure determination to confirm this. Several of the proposed molecular formulae do not have a match in databases (Scifinder, FoodDB, etc.) suggesting that it is very likely that some of these metabolites will be novel biomarkers for methane mitigation.
4 Conclusion
In conclusion, metabolomic and methanogenic profiles of the biserrula core collection are very congruent. We suggest that metabolites discovered in this study and most likely the ursolic or oleanolic terpene glycosides affect the methanogenic potential in the rumen and the novel ones can be potentially used for vaccine manufacturing in the future. Further mass spectrometry analysis of samples with CH4 output measurements in the field will be needed to confirm the relevance and true effects of the discovered metabolic markers. The results obtained from all datasets suggest that the Eritrean accessions likely belong to a different taxon within the species B. pelecinus with potentially different agronomic traits. Finally, crossing the low and high methanogenic accessions of biserrula investigated in this study will lead to materials and populations for gene discovery of CH4 output through transcriptomics and reverse genomics tools and techniques in the near future.
References
Baker SK, Gnanasampanthan G, Purser DB, Hoskinson RM (2004) Immunogenic preparation and method for improving the productivity of ruminant animals. Commonwealth Scientific and Industrial Research Organization (CSIRO), U.S. Patent 6,689,354
Banik BK, Durmic Z, Erskine W, Nichols P, Ghamkhar K, Vercoe P (2013) Variability of in vitro ruminal fermentation and methanogenic potential in the pasture legume biserrula (Biserrula pelecinus L.). Crop Pasture Sci 64(4):409–416. https://doi.org/10.1071/CP13073
Banik BK, Durmic Z, Erskine W, Revell CK, Vadhanabhuti J, McSweeney CS, Padmanabha J, Flematti GR, Algreiby AA, Vercoe PE (2016) Bioactive fractions from the pasture legume Biserrula pelecinus L. have an anti-methanogenic effect against key rumen methanogens. Anaerobe 39:173–182. https://doi.org/10.1016/j.anaerobe.2016.04.004
Bayaru E, Kanda S, Kamada T, Itabashi H, Andoh S, Nishida T, Ishida M, Itoh T, Nagara K, Isobe Y (2001) Effect of fumaric acid on methane production, rumen fermentation and digestibility of cattle fed roughage alone. Anim Sci J 72(2):139–146. https://doi.org/10.2508/chikusan.72.139
Bodas R, López S, Fernandez M, García-González R, Rodríguez A, Wallace R, González J (2008) In vitro screening of the potential of numerous plant species as antimethanogenic feed additives for ruminants. Anim Feed Sci Technol 145(1):245–258. https://doi.org/10.1016/j.anifeedsci.2007.04.015
Ghamkhar K, Revell C, Erskine W (2012) Biserrula pelecinus L.—genetic diversity in a promising pasture legume for the future. Crop Pasture Sci 63(9):833–839. https://doi.org/10.1071/CP12126
Ghamkhar K, Rochfort S, Banik B, Revell C (2016) Metabolomics for methane mitigation: a case study in Biserrula (Biserrula pelecinus L.). In: Roldán-Ruiz I, Baert J, Reheul D (eds) Breeding in a world of scarcity: proceedings of the 2015 Meeting of the Section “Forage Crops and Amenity Grasses” of Eucarpia. Springer International Publishing, Cham, pp 271–275
Gworgwor Z, Mbahi T, Yakubu B (2006) Environmental implications of methane production by ruminants: a review. J Sustain Dev Agric Environ 2:1
He X, Liu RH (2007) Triterpenoids isolated from apple peels have potent antiproliferative activity and may be partially responsible for apple’s anticancer activity. J Agric Food Chem 55(11):4366–4370. https://doi.org/10.1021/jf063563o
Heinzen H, de Vries JX, Moyna P, Remberg G, Martinez R, Tietze LF (1996) Mass spectrometry of labelled triterpenoids: thermospray and electron impact ionization analysis. Phytochem Anal 7(5):237–244. https://doi.org/10.1002/(SICI)1099-1565(199609)7:5<237::AID-PCA310>3.0.CO;2-M
Helander IM, Alakomi H-L, Latva-Kala K, Mattila-Sandholm T, Pol I, Smid EJ, Gorris LG, von Wright A (1998) Characterization of the action of selected essential oil components on Gram-negative bacteria. J Agric Food Chem 46(9):3590–3595. https://doi.org/10.1021/jf980154m
Holter J, Young A (1992) Methane prediction in dry and lactating Holstein cows. J Dairy Sci 75(8):2165–2175. https://doi.org/10.3168/jds.S0022-0302(92)77976-4
Joblin K (1999) Ruminal acetogens and their potential to lower ruminant methane emissions. Crop Pasture Sci 50(8):1307–1314. https://doi.org/10.1071/AR99004
Johnson KA, Johnson DE (1995) Methane emissions from cattle. J Anim Sci 73(8):2483–2492. https://doi.org/10.2527/1995.7382483x
Lee SS, Hsu J-T, Mantovani HC, Russell JB (2002) The effect of bovicin HC5, a bacteriocin from Streptococcus bovis HC5, on ruminal methane production in vitro. FEMS Microbiol Lett 217(1):51–55. https://doi.org/10.1111/j.1574-6968.2002.tb11455.x
Martin C, Morgavi D, Doreau M (2010) Methane mitigation in ruminants: from microbe to the farm scale. Animal 4(03):351–365. https://doi.org/10.1017/S1751731109990620
Mitsumori M, Sun W (2008) Control of rumen microbial fermentation for mitigating methane emissions from the rumen. Asian Australas J Anim Sci 21(1):144–154
Moss AR, Jouany JP, Newbold J (2000) Methane production by ruminants: its contribution to global warming. Ann Zootech 49(3):231–253. https://doi.org/10.1051/animres:2000119
Neto CC (2007) Cranberry and its phytochemicals: a review of in vitro anticancer studies. J Nutr 137(1):186S–193S
Patra AK, Saxena J (2009) Dietary phytochemicals as rumen modifiers: a review of the effects on microbial populations. Antonie Van Leeuwenhoek 96(4):363–375. https://doi.org/10.1007/s10482-009-9364-1
Patra A, Kamra D, Agarwal N (2006) Effect of plant extracts on in vitro methanogenesis, enzyme activities and fermentation of feed in rumen liquor of buffalo. Anim Feed Sci Technol 128(3):276–291. https://doi.org/10.1016/j.anifeedsci.2005.11.001
Pollier J, Goossens A (2012) Oleanolic acid. Phytochemistry 77:10–15. https://doi.org/10.1016/j.phytochem.2011.12.022
Rochfort S, Ezernieks V, Mele P, Kitching M (2015) NMR metabolomics for soil analysis provide complementary, orthogonal data to MIR and traditional soil chemistry approaches—a land use study. Magn Reson Chem 53(9):719–725. https://doi.org/10.1002/mrc.4187
Sejian V, Lal R, Lakritz J, Ezeji T (2011) Measurement and prediction of enteric methane emission. Int J Biometeorol 55(1):1–16. https://doi.org/10.1007/s00484-010-0356-7
Swinny E, Revell C, Campbell N, Spadek E, Russo C (2015) In search of photosensitising compounds in the annual forage legume Biserrula pelecinus L. Crop Pasture Sci 66(11):1161–1166. https://doi.org/10.1071/CP14342
Szakiel A, Ruszkowski D, Janiszowska W (2005) Saponins in Calendula officinalis L.—structure, biosynthesis, transport and biological activity. Phytochem Rev 4(2–3):151–158. https://doi.org/10.1007/s11101-005-4053-9
Ulyatt M, Lassey K, Shelton I, Walker C (2002) Methane emission from dairy cows and wether sheep fed subtropical grass-dominant pastures in midsummer in New Zealand. N Z J Agric Res 45(4):227–234. https://doi.org/10.1080/00288233.2002.9513513
Wallace RJ, McEwan NR, McIntosh FM, Teferedegne B, Newbold CJ (2002) Natural products as manipulators of rumen fermentation. Asian Australas J Anim Sci 15(10):1458–1468
Woodward SL, Waghorn GC, Lassey KR, Laboyrie PG (2002) Does feeding sulla (Hedysarum coronarium) reduce methane emissions from dairy cows? Proceedings-New Zealand Society of Animal Production 62:227–230
Xia Y-G, Liang J, Li G-Y, Yang B-Y, Kuang H-X (2015) Analysis of oligosaccharide sequences of trace Caulophyllum robustum saponins by direct infusion multiple-stage tandem mass spectrometry. J Pharm Biomed Anal 112:106–115. https://doi.org/10.1016/j.jpba.2015.04.024
Acknowledgements
The authors would like to thank Vilnis Ezernieks of the Department of Economic Development, Victoria, for MS lab experiments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Ghamkhar, K., Rochfort, S., Banik, B.K. et al. Candidate metabolites for methane mitigation in the forage legume biserrula. Agron. Sustain. Dev. 38, 30 (2018). https://doi.org/10.1007/s13593-018-0510-x
Accepted:
Published:
DOI: https://doi.org/10.1007/s13593-018-0510-x