Abstract
Phoretic mites associated with bumblebees are considered commensals that represent little or no risk for their hosts. The aim of the present study was to assess the potential role of the phoretic mites Pneumolaelaps longanalis and Parasitellus fucorum, as carriers of parasites that are known to affect bees. Mites were extracted from bumblebees dating between 1945 and 1981 which belong to museum collections, and from a freshly collected queen. The DNA was extracted and amplified, and the final products were sequenced and analyzed. Parasite’s DNA was found in all samples. Ascosphaera spp., Crithidia bombi, Lotmaria passim, and Nosema ceranae were present in both mite species. Moreover, DNA from Apis mellifera filamentous virus (AmFV), Apicystis bombi, Ascosphaera apis, Nosema apis, and Nosema bombi was found in P. longanalis. This study represents the oldest report of parasite’s DNA in bumblebee phoretic mites, highlighting the relevance of museum samples for historical studies in parasitology.
Similar content being viewed by others
1 Introduction
Bees are major angiosperm pollinators, providing a key service to crop production estimated in more than a hundred billion Euros per year (Gallai et al. 2009; Klein et al. 2013). However, and despite their importance, it is known that both domestic and wild bee populations are threatened worldwide (Freitas et al. 2009; Neumann and Carreck 2010; Potts et al. 2010; Maggi et al. 2016). These bees provide pollination services in both natural and managed environments (Goulson et al. 2008); therefore, their reduction in abundance and distribution range imply an important ecological and economic detriment (Biesmeijer et al. 2006). The main proposed drivers for these phenomena include pathogens and other parasites, interaction with exotic species, agrochemicals, habitat fragmentation, and climate change (Goulson 2010). In most cases, a combination of such stressors, rather than a particular one, leads to the decline of several wild bee populations and severe honeybee colony losses (Kluser and Peduzzi 2007; Brown and Paxton 2009; Potts et al. 2010; Cameron et al. 2011; Goulson et al. 2015; Meeus et al. 2018).
The western honeybee, Apis mellifera, and the buff-tailed bumblebee, Bombus terrestris, are by far the most studied bee species. Many pathogens were found affecting these bees, like fungi, mites, viruses, and protists, which are the most frequent causative agents of disease (Bailey and Ball 1991; Ruiz-González and Brown 2006; Meeus et al. 2010). Recently, it was reported that pathogen spillover from managed to wild bee populations could have potential negative consequences for both crop pollination and conservation of native bees (Blitzer et al. 2012).
Mites and bees are closely associated exhibiting broad range of interaction from phoresy to parasitism (Chmielewski 1971; Baker 2000; Maggi et al. 2011; Revainera et al. 2014). Particularly important is the case of Varroa destructor, considered as one of the main factors contributing to honeybee colony losses around the world (Steinhauer et al. 2018). The mite feeds on fat tissue of both larvae and adult bees (Ramsey et al. 2018), acting at the same time as vector of many viruses, like black queen cell virus (BQCV), deformed wing virus (DWV), acute bee paralysis virus (ABPV), and Israeli acute paralysis virus (IAPV) (Martin et al. 2012).
Phoretic mites associated with bumblebees use the insect as a transport to reach a nest in order to obtain nutritional supply such as pollen, nectar, comb material, nematodes, and microorganisms (Walter et al. 2002; Cordeiro et al. 2011). This association could benefit both parts of the bee-mite system, where mites reach a secure place for feeding and reproduction, meanwhile bees have their nests cleaned by mites (Schwarz and Huck 1997). On the other hand, Athias-Binche (1994) proposed that mites represent a disturbance for bees, since they could hamper the flight when are present in high loads, and also to interrupt the communication, when they touch the antennae or mouthparts. In addition, mites could be able to induce starvation in the bee colony when they feed on nest reservoirs during rainy seasons (Haq et al. 2011). Food reservoirs provide nutrients and a suitable habitat for fungi inside the nests, and mites like those of the genera Parasitellus, Scutacarus, and Tyrophagus (Hughes 1976; Hubert et al. 2003; Marjorie 2011) feed on these fungi and are able to transport their spores to new areas. However, there are no conclusive results providing that mites constitute a relevant threat for bumblebees. Phoretic mites can be transferred between bees of the same species switching from (i) workers to queens, probably detecting queen features like queen pheromones (Huck et al. 1998), (ii) males to queens during copulation (O’Connor 1982), and (iii) among bees from the same or different species during foraging (Schwarz and Huck 1997). Given the lack of specificity of these mites (Maggi et al. 2011; Revainera et al. 2014; Revainera et al. 2019), they could reach nests of different bee species carrying potential pathogens.
The mesostigmatid mite species Parasitellus fucorum (Parasitidae) and Pneumolaelaps longanalis (Laelapidae) are kleptoparasites that present a phoretic deutonymph and a life cycle closely associated with bumblebee nests (Royce and Krantz 1989; Koulianos and Schwarz 1999). The adult mites and deutonymphs feed on the pollenkit and nectar coating that bees apply to the pollen grains, leaving them with a pale and translucent aspect, with no other damage, although P. longanalis is also able to break the pollen grains to obtain more nutrients (Royce and Krantz 1989; Koulianos and Schwarz 1999). The high proportion of bumblebees observed with phoretic P. longanalis and P. fucorum (prevalence), the high number of these mites on each bumblebee (intensity), and their big size compared with other common phoretic mites such as Kuzinia and Scutacarus are characteristics that make them good targets for parasitological studies.
The aims of this study were to (i) look for parasites in P. longanalis and P. fucorum, isolated from a current, field-collected sample as well as from museum collections and (ii) evaluate the detection efficiency of different primer pairs for parasite’s DNA from collection samples.
2 Materials and methods
2.1 Sample collection
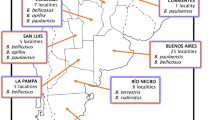
The phoretic mite species P. longanalis and P. fucorum were extracted from a Bombus pauloensis foraging queen in 2018 and identified using taxonomic keys (Hunter 1966; Hyatt 1980). Additionally, twelve specimens of B. pauloensis, Bombus dahlbomii, Bombus morio, and Bombus opifex deposited in La Plata Museum and dated from 1947 to 1981 were examined, from which several specimens of P. longanalis were extracted. Host species, number of extracted mites, locality, and date of collection are detailed in Table I and Figure 1.
2.2 DNA extraction and qPCR control
Total genomic DNA was extracted from pool homogenate of mites (Table I) using the High Pure PCR Template Preparation Kit (Roche Diagnostics). To verify the success of DNA extraction from pools and lack of inhibition in the PCR, DNA amplifications of a 138-bp PCR product of 18S ribosomal (rDNA) with primers 18SEu F and 18SEu R (Table II) were performed (Fajardo et al. 2008).
The cycling program consisted of an initial denaturation of 2 min at 95 °C, and 40 cycles of 94 °C (20″), 52 °C (20″), and 72 °C (30″). After amplification, a melting curve analysis was performed, which resulted in single product-specific melting curve. Those samples with 18S rDNA Ct (cycle threshold) values < 30 were considered suitable.
2.3 Pathogen detection through qPCR
In order to study the presence of pathogen DNA in mites, we used primer sets selected from bibliography. We screened for Apis mellifera filamentous virus (AmFV), Apicystis bombi (Apicomplexa: Neogregarinorida), Crithidia bombi and Lotmaria passim (Euglenozoa: Trypanosomatida), Nosema ceranae, Nosema bombi, and Nosema apis (Fungi: Microsporidia), and Ascosphaera spp. (Fungi: Ascomycota), pathogens that are known to infest honeybees and bumblebees, with negative effects on their health (Macfarlane et al. 1995; Genersch et al. 2009). The cycling program consisted of an initial denaturation of 2 min at 95 °C, and 40 cycles of 95 °C (20″), annealing temperature (20″), and 72 °C (30″). Samples with Ct values < 30 were considered positive. To search for AmFV, L. passim, and N. bombi, two sets of primers (of different product length) each were used. The specific dissociation temperatures of each searched pathogen with each pair of primers are detailed in Table II. The reaction efficiency for each primer pair was calculated, in all cases being between 95 and 105%. During the validation process, the PCR products were run on agarose gels to check the size of the PCR products. All qPCR were carried out in a Rotor Gene thermocycler (Qiagen, Hilden, Germany) in a final volume of 20 μl using EvaGreen as an intercalating fluorescent dye (KAPA FAST, Biosystems, Woburn, USA). In each qPCR run, positive, negative, and non-template controls were added. As positive controls, DNA extracted from pathogen isolates was used in all cases, except for AmFV, in which a positive honeybee sample was used. Negative controls consisted in DNA from samples in which the absence of the pathogen’s DNA was confirmed previously.
To verify the specificity of the selected primers, amplified DNA fragments were purified and directly sequenced (ABI 3500 Genetic Analyzer, Applied Biosystems, Foster City, CA, USA). The sequence similarities were determined by the Basic Local Alignment Search Tool (BLAST, NCBI).
3 Results
A total of 88 mites were extracted and pooled from 13 bees. As DNA extraction control, a 138-bp PCR product of 18S ribosomal (rDNA) was amplified in all the analyzed samples, showing that they were suitable for further study.
Parasite’s DNA was detected in all samples. Both mite species, P. longanalis and P. fucorum, were positive for C. bombi, L. passim, N. ceranae, and Ascosphaera spp. Additionally, P. longanalis mites showed the presence of DNA of A. bombi, AmFV, N. bombi, N. apis, and A. apis (Table III).
AmFV was present in all the collection samples, with the oldest one from 1947, and also present in a pool of mites collected in 2018. Apicystis bombi was only detected in a sample from 1951. DNA from an undetermined species of Ascosphaera was detected in the two new samples and in one from 1981, whereas A. apis was found in a sample from 1945. The trypanosomatids were detected in pools from 2018 as well as in the ones taken from museum collections, with the oldest sample (of both species) dated in 1948. The three microsporidia were also found; both N. ceranae and N. bombi were detected in the two current pools of mites, while N. apis was detected only in one. Their oldest records were from 1947, 1948, and 1951 for N. ceranae, N. bombi, and N. apis, respectively.
Regarding analysis of AmFV, L. passim, and N. bombi, the set of primers that amplified a shorter sequence improved the detection capability of these pathogens in all cases. Detection of AmFV increased from 28% with AmFV551 F/AmFV551 R set to 93% with AmFV97 F/AmFV97 R. Lotmaria passim and N. bombi were only detected using the shorter set (50% with qCrF153/qCrR15, and 28% with Bombicar101 F/Bombicar101 R, respectively).
The presence of the analyzed parasites was confirmed by the sequencing of the obtained PCR products. Lotmaria passim sequence obtained with primers qCr from mite p1 (GenBank accession no. MK860770) and mite p8 (GenBank accession no. MK922078) showed 97% and 99% homology with L. passim from Uruguay (GenBank accession no. KX953204.1). AmFV sequence amplified with primers AmFV97 from mite p4 and with primers AmFV551 from mite p12 (GenBank accession no. MK941661) showed both 100% and 94% of homology with AMFV partial genome from Switzerland (GenBank accession no. KR819915.1), respectively. Crithidia bombi sequence from mite p10 (GenBank accession no. MK929304) showed 95% homology with C. bombi from India (GenBank accession no. KX151692.1). Nosema ceranae sequence from mites p10 (GenBank accession no. MK918502) and p2 (GenBank accession no. MK844210) showed 98% and 99% of homology, respectively, with N. ceranae from Lithuania (GenBank accession no. JQ639307.1) and Mexico (GenBank accession no. HM802210.1). Nosema bombi sequence from mites p2 (GenBank accession no. MK922077) and ab5 (GenBank accession no. MK929303) showed 98% and 97% of homology with N. bombi from Thailand (GenBank accession no. MF776551.1), respectively. Nosema apis sequence from mite p8 (GenBank accession no. MK875322) showed 98.8% of homology with N. apis from Poland (GenBank accession no. KC707997.1). In the case of A. bombi, Ascosphaera sp., and A. apis, the amplified fragments of DNA were scarce in quantity, not being adequate for sequencing, thus impeding any further determinations.
4 Discussion
The finding of parasite’s DNA in P. longanalis from samples dated between 1945 and 1981 represents a new paradigm in the historical distribution of the infestation phenomenon. Our results suggest that almost all detected pathogens have been present in native bees for more than 50 years whereas most of them were described decades after the dates recorded in this report. Ascosphaera apis, the causative agent of the Chalkbrood disease of A. mellifera, has been described at the beginnings of the twentieth century (Maassen 1913), and their spores were also recently isolated from Bombus spp. (Maxfield-Taylor et al. 2015), but its presence in Argentina has not been detected prior to 1978 (Rossi and Carranza 1980). Our results suggest that A. apis dates back at least to the 1940s.
Moreover, after a survey in South America (Plischuk and Lange 2009), it has been proposed that A. bombi, C. bombi, or (more recently) N. bombi could have been introduced by alien bees and then transferred to native ones (Arbetman et al. 2013; Plischuk et al. 2017); however, temporarily space incompatible findings have suggested some weaknesses in this hypothesis (Gamboa et al. 2015; Nunes-Silva et al. 2016). Throughout our contribution, we suggest that these three species were in fact present in the region long before the introduction of B. ruderatus in 1982 and B. terrestris in 1998.
The microsporidium N. ceranae is a highly pathogenic fungus that was proposed as one of the main factors involved in the worldwide decline of wild bee populations and honeybee colony losses. It was suspected that this parasite shifted from Apis cerana to A. mellifera sometime before 1990 (Invernizzi et al. 2009). Additionally, the first report of N. ceranae in South American native bumblebees dated from 2005 (Plischuk et al. 2009). However, in this study, we reported its presence in mites closely associated with bumblebees, the flowers that they visit, and their nests, from at least 1945. In this line of evidence, the spread of N. ceranae to other bee species could have occurred many decades earlier than suspected.
The presence of L. passim in mites is also intriguing. This flagellate has been found only in A. mellifera gut, so its finding in bumblebee phoretic mites is difficult to explain. Possibly, as Durrer and Schmid-Hempel (1994), Ruiz-González and Brown (2006), and Graystock et al. (2015) suggested, there is an exchange of pathogens by sharing flowers during pollination, meanwhile their ectoparasites would be more frequent than is currently supposed.
Although some of the parasites found in the present study have a slight effect on honeybees’ health, others are highly virulent (Bailey and Ball 1991), and then their transmission via phoretic mites to native bee species could represent a problem, especially those of high prevalence. Despite the uncertainty about if the transported parasites were in an infective form, those that produce resistance structures (such as Ascosphaera, Nosema, or A. bombi) could be more prone to remain active until they reach their definitive hosts. These organisms are also easily dispersed, thus even if in the laboratory the hygiene protocols followed ensure the lack of contamination, cross-contamination could occur in the museum.
In this work, we compared three primer sets for the detection of L. passim, N. bombi, and AmFV DNA, finding that the corresponding pair of the smallest PCR product showed the best detection capability. Probably, primers that amplified larger products were not able to generate PCR products due to DNA degradation. When a PCR-based method is used in routine analysis, a DNA extraction control could indicate false-negative results (Hoofar et al. 2003; Burd 2010) and is critical for analyzing field-collected samples. Sample preservation in the field or in museums to ensure DNA stability is not always manageable; thus, there are many cases where samples are not correctly stored to guarantee such stability. In the particular case of field-sampled bees and their associated mites, there are various factors that can inhibit PCR, for instance the presence of pollen or even the bee’s compound eyes (Boncristiani et al. 2011; Lalhmangaihi et al. 2014). Here, we used the amplification of a 138-bp PCR product as an internal control to verify DNA quality. It should be noted that the size of the internal control is also an important issue. In this work, all samples were positive for the internal control, but some of them only amplified N. bombi, AmFV, and L. passim DNA with the smallest primer set. The size of the internal control should be similar to the size of the PCR product for the detection of the specific parasite.
This study represents the first detection of certain bee pathogens in phoretic mites. Although contamination in the museum, especially by spores, cannot be ruled out, the presence of parasite’s DNA in such high proportions should not be dismissed, since transmission to new hosts by mites could represent a threat to several bee species. Moreover, co-occurrence of two or more parasites in mite samples could be detrimental for the bees, as noted by (Alizon and Lion 2011), for N. ceranae and L. passim. Here, we show that phoretic mite prospection could be considered as a valuable method for colony health estimations even for ancient bee populations, helping to reconstruct the historical dynamics of certain infections. Thus, the study of museum collections opens new frontiers within the study of historical parasitology in bees. We suggest that further studies should be focused on the estimation of parasite virulence in mites, for exploring the role that such mites could play in wild bee population declines.
References
Alizon, S., Lion, S. (2011) Within-host parasite cooperation and the evolution of virulence. Proc. R. Soc. B, 278, 3738–3747.
Arbetman, M.P., Meeus, I., Morales, C.L., Aizen, M.A., Smagghe, G. (2013) Alien parasite hitchhikes to Patagonia on invasive bumblebee. Biol. Invasions., 15, 489–494.
Arismendi, N., Bruna, A., Zapata, N., & Vargas, M. (2016) PCR-specific detection of recently described Lotmaria passim (Trypanosomatidae) in Chilean apiaries. J Invertebr. Pathol., 134, 1–5.
Athias-Binche, F. (1994) La Phoresie chez les acariens: aspects adaptatifs et évolutifs. Editions du Castillet. Perpignan, 179 pp.
Bailey, L., Ball, B.V. (1991) Honeybee pathology. Academic Press, London. 193 pp.
Baker, R.A. (2000). Mites and bees: an historical review. Pszczeln. Zesz. Nauk., 44(2), 109–113.
Biesmeijer, J.C., Roberts, S.P.M., Reemer, M., Ohlemuller, R., Edwards, M., Peeters, T., Schaffers, A.P., Potts, S.G., Kleukers, et al. (2006) Parallel declines in pollinators and insect pollinated plants in Britain and the Netherlands. Science, 313, 351–354.
Blitzer, E.J., Dormann, C.F., Holzschuh, A., Klein, A.M., Rand, T.A., Tscharntke, T. (2012) Spillover of functionally important organisms between managed and natural habitats. Agric. Ecosyst. Environ., 146(1), 34–43.
Boncristiani, H., Li, J.L., Evans, J.D., Pettis, J., Chen, Y.P. (2011) Scientific note on PCR inhibitors in the compound eyes of honeybees, Apis mellifera. Apidologie, 42, 457–460.
Brown, M.J., Paxton, R.J. (2009) The conservation of bees: a global perspective. Apidologie, 40(3), 410–416.
Burd, E.M. (2010) Validation of laboratory-developed molecular assays for infectious diseases. Clin. Microbiol. Rev., 23, 550–576.
Cameron, S.A., Lozier, J.D., Strange, J.P., Koch, J.B., Cordes, N., Solter, L.F., Griswold, T.L. (2011) Patterns of widespread decline in North American bumble bees. Proc. Natl. Acad. Sci. 108, 662–667.
Chmielewski, W. (1971) The mites (Acarina) found on bumblebees (Bombus Latr.) and in their nests. Ekologia Polska, 19, 57–71.
Cordeiro, G.D., Taniguchi, M., Flechtmann, C.H.W., Alves-dos-Santos, I. (2011) Phoretic mites (Acari: Chaetodactylidae) associated with the solitary bee Tetrapedia diversipes (Apidae: Tetrapediini). Apidologie, 42(2), 128–139.
Durrer, S., Schmid-Hempel, P. (1994) Shared use of flowers leads to horizontal pathogen transmission. Proc. R. Soc. B., 258(1353), 299–302.
Fajardo, V., Gonzalez, I., Martin, I., Rojas, M., Hernandez, P.E., Garcia, T., Martin, R. (2008) Real–time PCR for detection and quantification of red deer (Cervus elaphus), fallow deer (Dama dama), and roe deer (Capreolus capreolus) in meat mixtures. Meat. Sci., 79(2), 289–298.
Freitas, B.M., Imperatriz-Fonseca, V.L., Medina, L.M., Kleinert, A.D.M.P., Galetto, L., Nates-Parra, G., Quezada-Euán, J.J.G. (2009) Diversity, threats and conservation of native bees in the Neotropics. Apidologie, 40(3), 332–346.
Gallai, N., Salles, J.M., Settele, J., Vaissière, B.E. (2009) Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol. Econ., 68, 810–821.
Gamboa, V., Ravoet, J., Brunain, M., Smagghe, G., Meeus, I., Figueroa, J., Riaño, D., de Graaf, D.C. (2015) Bee pathogens found in Bombus atratus from Colombia: a case study. J. Invertebr. Pathol., 129, 36–39.
Genersch, E., Evans, J.D., Fries, I. (2009) Honey bee disease overview. J. Invertebr. Pathol. 103, S2-S4.
Goulson, D. (2010) Bumblebees: behaviour, ecology, and conservation. Oxford Univ. Press, Oxford.
Goulson, D., Lye, G.C., Darvill, B. (2008) Decline and conservation of bumblebees. Ann. Rev. Entomol., 53, 191–208.
Goulson, D., Nicholls, E., Botías, C., Rotheray, E.L. (2015) Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science, 347(6229), 1255957.
Graystock, P., Goulson D., Hughes, W.O.H. (2015) Parasites in bloom: flowers aid dispersal and transmission of pollinator parasites within and between bee species. Proc. R. Soc. B, 282, 20151371.
Haq, M.A., Sumangala, K., Ramani, N. (2011) Mites associated with concealed and open nests of Apis cerana indica in Kerala, South India. In Trends in Acarology. pp. 371-372. Springer, Netherlands.
Hartmann, U., Forsgren, E., Charrière, J.D., Neumann, P., Gauthier, L. (2015) Dynamics of Apis mellifera filamentous virus (AmFV) infections in honey bees and relationships with other parasites. Viruses, 7(5), 2654–2667.
Hoofar, J., Cook, N., Malorny, B., Wagner, M., De Medici, D., Abdulmawjood, A., and Fach, P. (2003) Making internal amplification control mandatory for diagnostic PCR. J. Clinical. Microbiol., 41, 5835.
Huang, W.F., Skyrm, K., Ruiter, R., Solter, L. (2015) Disease management in commercial bumble bee mass rearing, using production methods, multiplex PCR detection techniques, and regulatory assessment. J. Apicult. Res., 54(5), 516–524.
Hubert, J., Stejskal, V., Kubátová, A., Munzbergová, Z., Váňová, M., Žd’árková, E. (2003) Mites as selective fungal carriers in stored grain habitats. Exp. App. Acarol., 29 (1), 69–87.
Huck, K., Schwarz, M., Schmid-Hempel, P. (1998) Host choice in the phoretic mite Parasitellus fucorum (Mesostogmata: Parasitidae): which bumblebee caste is the best? Oecologia, 115, 385–390.
Hughes, A.M. (1976) The mites of stored food and houses, 2nd ed. Ministry of Agriculture, Fisheries and Food, Technical Bulletin 9. Her Majesty’s Stationery Office, London, 400 pp.
Hunter, P.E. (1966) The genus Pneumolaelaps with description of three new species (Acarina: Laelaptidae). J. Kansas Entomol. Soc., 39, 357–369.
Hyatt, K.H. (1980) Mites of the subfamily Parasitinae (Mesostigmata: Parasitidae) in the British Isles. Bull. br. Mus. nat. Hist. Zool., 38, 237–378.
Invernizzi, C., Abud, C., Tomasco, I. H., Harriet, J., Ramallo, G., Campá, J., Katz, H., Gardiol, G., Mendoza, Y. (2009) Presence of Nosema ceranae in honeybees (Apis mellifera) in Uruguay. J. Invertebr. Pathol., 101(2), 150–153.
James, R.R., Skinner, J.S. (2005) PCR diagnostic methods for Ascosphaera infections in bees. J. Invertebr. Pathol., 90(2), 98–103.
Klee, J., Tay, W.T., Paxton, R.J. (2006) Specific and sensitive detection of Nosema bombi (Microsporidia: Nosematidae) in bumble bees (Bombus spp.; Hymenoptera: Apidae) by PCR of partial rRNA gene sequences. J. Invertebr. Pathol. 91(2), 98-104.
Klein, A.M., Steffan-Dewenter, I., Tscharntke, T. (2013) Fruit set of highland coffee increases with the diversity of pollinating bees. Proc. R. Soc. B, 270, 955–961.
Kluser, S., Peduzzi, P. (2007) Global pollinator decline: a literature review.
Koulianos, S., Schwarz, H.H. (1999) Reproduction, development and diet of Parasitellus fucorum (Mesostigmata: Parasitidae), a mite associated with bumblebees (Hymenoptera: Apidae). J. Zool., 248, 267–269.
Lalhmangaihi, R., Ghatak, S., Laha, R., Gurusubramanian, G., Kumar, N.S. (2014) Protocol for optimal quality and quantity pollen DNA isolation from honey samples. J. Biomol. Tech., 25, 92–95.
Maassen, A. (1913). Weitere Mitteilungen uber der seuchenhaften Brutkrankheiten der Bienen. Mitteilungen aus der Kaiserlichen Biologischen Anstalt fur Land-und Forstwirtscshaft, 14, 48–58.
Macfarlane, R. P., Lipa, J. J., Liu, H. J. (1995) Bumble bee pathogens and internal enemies. Bee World, 76(3), 130–148.
Maggi, M., Lucia, M., Abrahamovich, A. H. (2011) Study of the acarofauna of native bumblebee species (Bombus) from Argentina. Apidologie, 42(3), 280–292.
Maggi, M., Antúnez, K., Invernizzi, C., Aldea, P., Vargas, M., Negri, P., Brasesco, C., De Jong, D., Message, D., Weinstein Teixeira, E., Principal, J., Barrios, C., Ruffinengo, S., Rodríguez Da Silva, R., Eguaras, M. (2016) Honeybee health in South America. Apidologie, 47(6), 835–854.
Marjorie, A.H. (2011) Agricultural acarology. CRC Press, New York. 410 pps.
Martin, S.J., Highfield, A.C., Brettell, L., Villalobos, E.M., Budge, G.E., Powell, M., Nikaido, S., Schroeder, D.C. (2012) Global honeybee viral landscape altered by a parasitic mite. Science, 336(6086), 1304–1306.
Martín-Hernández, R., Meana, A., Prieto, L., Salvador, A. M., Garrido-Bailón, E., Higes, M. (2007) Outcome of colonization of Apis mellifera by Nosema ceranae. App. Environ. Microbiol., 73(20), 6331–6338.
Maxfield-Taylor, S.A., Mujic, A.B., Rao, S. (2015) First detection of the larval chalkbrood disease pathogen Ascosphaera apis (Ascomycota: Eurotiomycetes: Ascosphaerales) in adult bumblebees. PLoS One, 10(4), 1–11.
Meeus, I., De Graaf, D. C., Jans, K., Smagghe, G. (2010). Multiplex PCR detection of slowly-evolving trypanosomatids and neogregarines in bumblebees using broad-range primers. J. App. Microbiol., 109(1), 107–115.
Meeus, I., Pisman, M., Smagghe, G., Piot, N. (2018) Interaction effects of different drivers of wild bee decline and their influence on host-pathogen dynamics. Curr. Opi. Insect Sci., 26, 136–141.
Neumann, P., Carreck, N.L. (2010) Honey bee colony losses. J Apic Res, 49, 1–6.
Nunes-Silva, P., Piot, N., Meeus, I., Blochtein, B., Smagghe, G. (2016) Absence of Leishmaniinae and Nosematidae in stingless bees. Sci. Rep., 6, 32547.
O’Connor, B.M. (1982) Evolutionary ecology of astigmatid mites. Annu. Rev. Entomol. 27, 385–409.
Plischuk, S., Lange, C.E. (2009) Invasive Bombus terrestris (Hymenoptera: Apidae) parasitized by a flagellate (Euglenozoa: Kinetoplastea) and a neogregarine (Apicomplexa: Neogregarinorida). J. Invertebr. Pathol., 102, 261–263.
Plischuk, S., Martín-Hernández, R., Prieto, L., Lucía, M., Botías, C., Meana, A., Higes, M. (2009) South American native bumblebees (Hymenoptera: Apidae) infected by Nosema ceranae (Microsporidia), an emerging pathogen of honeybees (Apis mellifera). Environ. Microbiol. Rep., 1(2), 131–135.
Plischuk, S., Antúnez, K., Haramboure, M., Minardi, G. M., Lange, C. E. (2017) Long-term prevalence of the protists Crithidia bombi and Apicystis bombi and detection of the microsporidium Nosema bombi in invasive bumblebees. Environ. Microbiol. Rep., 9(2), 169–173.
Potts, S.G., Roberts, S.P., Dean, R., Marris, G., Brown, M.A., Jones, H.R., Neumann, P., Settele, J. (2010) Declines of managed honeybees and beekeepers in Europe. J. Apicult. Res., 49, 15–22.
Ramsey, S., Gulbronson, C.J., Mowery, J., Ochoa, R., Bauchan, G. (2018) A multi-microscopy approach to discover the feeding site and host tissue consumed by Varroa destructor on host honeybees. Microsc. Microanal., 24,1258–1259.
Revainera, P., Lucia, M., Abrahamovich, A.H., Maggi, M. (2014). Spatial aggregation of phoretic mites on Bombus atratus and Bombus opifex (Hymenoptera: Apidae) in Argentina. Apidologie, 45(5), 579–589.
Revainera, P.D., Salvarrey, S., Santos, E., Arbulo, N., Invernizzi, C., Plischuk, S., Abrahamovich, A.H., Maggi, M.D. (2019) Phoretic mites associated to Bombus pauloensis and Bombus bellicosus (Hymenoptera: Apidae) from Uruguay. J. Apicult. Res., 58 (3), 455–462.
Rossi, C.O., Carranza, M.R. (1980) Momificación de larvas de abeja (Apis mellifera L.) provocada por Ascosphaera apis. Rev. Fac. Agron. 56(1–2), 11-15.
Royce, L.A., Krantz, G.W. (1989) Observations on pollen processing by Pneumolaelaps longanalis (Acari, Laelapidae), a mite associate of bumblebees. Exp. App. Acarol., 7, 161–165.
Ruiz-González, M.X., Brown, M.J. (2006) Honey bee and bumblebee trypanosomatids: specificity and potential for transmission. Ecol. Entomol., 31(6), 616–622.
Runckel, C., Flenniken, M.L., Engel, J.C., Ruby, J.G., Ganem, D., Andino, R., de Risi, J.L. (2011). Temporal analysis of the honey bee microbiome reveals four novel viruses and seasonal prevalence of known viruses, Nosema, and Crithidia. PLoS One, 6, e20656.
Schwarz, H.H., Huck, K. (1997) Phoretic mites use flowers to transfer between foraging bumblebees. Insect. Soc., 44, 303–310.
Steinhauer, N., Kulhanek, K., Antunez, K., Human, H., Chantawannakul, P., Chauzat, M. P. (2018) Drivers of colony losses. Curr. Opi. Insect Sci., 26, 142–148.
Walter, D.E., Beard, J.J., Walker, K.L., Sparks, K. (2002) Of mites and bees: a review of mite–bee associations in Australia and a revision of Raymentia Womersley (Acari: Mesostigmata: Laelapidae), with the description of two new species of mites from Lasioglossum (Parasphecodes) spp. (Hymenoptera: Halictidae). Austral. J. Entomol. 41(2), 128-148.
Funding
This research was financially supported by FONCyT (PICT 2015-1045 granted to SQ, and PICT 2016-0289 granted to SP). The authors would like to thank the Institute of Analysis Fares Taie, CONICET, and the UNMdP for financial support.
Author information
Authors and Affiliations
Contributions
PR, SQ, and MM conceived the research. PR, GFL, and SQ performed experiments and analysis. All authors participated in the sampling and writing process and approved the final version.
Corresponding author
Additional information
Manuscript editor: Cedric Alaux
Les acariens phorétiques des bourdons d'Amérique du Sud ( Bombus spp.) comme porteurs de parasites : un apport historique.
Acariens phorétiques / Bourdons / ADN du parasite / Détection moléculaire / Parasitologie historique.
Phoretische Milben auf südamerikanischen Hummeln ( Bombus spp.) als Träger von Parasiten: eine historische Betrachtung.
Phoretische Milben / Hummeln / DNA von Parasiten / Molekularer Nachweis / historische Parasitologie.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Revainera, P.D., Quintana, S., Fernández de Landa, G. et al. Phoretic mites on South American bumblebees (Bombus spp.) as parasite carriers: a historical input. Apidologie 51, 455–464 (2020). https://doi.org/10.1007/s13592-020-00733-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13592-020-00733-w