Abstract
The stingless bee Tetragonilla collina Smith, 1857, is broadly distributed across Indochina. In this study, we use a combination of molecular and geometric morphometric analyses to quantify the genetic structure and diversity of the T. collina population of Thailand. We found striking regional differences in both mitochondrial haplotype frequencies and morphology. A Bayesian analyses of molecular diversity of the mitochondrial COI region revealed two clades, roughly divided into the population northeast of the Thai-Malay Peninsula (clade A) and the western and Thai-Malay Peninsula population (clade B). In addition, morphometric analysis showed that bees in clade A have significantly larger wings than bees from clade B. These results suggest that the T. collina population of Thailand is divided into two distinct populations. The spatial distributions seem to reflect contemporary ecological features such as annual flooding (bees of clade B are absent from areas subject to inundation), rather than past biogeography. Thus, T. collina differs from the honey bees Apis dorsata and A. cerana that show genetic differentiation north and south of the Isthmus of Kra, perhaps reflective of past separation during the Pleistocene when sea levels were much higher.
Similar content being viewed by others
1 Introduction
Geographical features in a landscape are often associated with abrupt transitions in species or subspecies distribution. These may be legacies of historical barriers to gene flow, caused by the geographical feature, or by a change in contemporary ecological processes that affect species distribution. The Isthmus of Kra on the Malay peninsula, between 10° 34′ N and 11° 24′ N, is a transition zone between seasonal rainforest and mixed moist deciduous forest (Hughes et al. 2003), and between Indochinese and Sundaic biota (Woodruff 2003). During the early Pliocene (5.5–4.5 million years ago), many plant and animal populations north of the Kra Isthmus were repeatedly separated from populations to the south by changing sea levels, leading to the creation of new species and subspecies (Woodruff 2003). Contemporary distributions often reflect these historical events. For example, three shrew species show a transition zone just south of Isthmus of Kra near Satun to Sai Suri (Roberts 2011). Additionally, the honey bees Apis cerana (Deowanish et al. 1996; Smith and Hagen 1996; Sihanuntavong et al. 1999; Sittipraneed et al. 2001; Warrit et al. 2006) and A. dorsata (Insuan et al. 2007) show significant genetic variation among geographical regions of Thailand, particularly between populations north and south of the Kra Isthmus (Sihanuntavong et al. 1999; Warrit et al. 2006). Interestingly, Varroa jacobsoni, a parasitic mite of A. cerana, also shows different haplotypes north and south of the Kra ecotone (Warrit et al. 2006).
The stingless bees are a large group of tropical eusocial bees of the tribe Meliponini (Kerr and Maule 1964). In contrast to honey bees, which reproduce and migrate by swarms that can travel many tens of kilometers from the natal nest (Koeniger and Koeniger 1980; Nakamura et al. 1991; Dyer and Seeley 1994; Itioka et al. 2001; Paar et al. 2004), stingless bee colonies reproduce by a gradual process of budding, which restricts the distance of daughter colonies from their natal nest to 100 m or so (Michener 1979; Inoue et al. 1984; van Veen and Sommeijer 2000; Roubik 2006; Francisco and Arias 2010). This reproductive behavior means that stingless bee populations tend to show much greater structure than honey bee populations (Francisco et al. 2008, 2014; Francisco and Arias 2010; Brito et al. 2014) and might be expected to reflect the vestiges of past biogeography even more strongly than honey bee populations. Tetragonilla collina Smith, 1857, is one of the most common and broadly distributed stingless bee species in the Indochina region (Sakagami and Khoo 1987) and is found throughout Thailand (Theeraapisakkun et al. 2010). It is therefore an ideal species to examine the hypothesis that the Isthmus of Kra is a transition zone for stingless bee subpopulations that were formerly separated north and south of the Isthmus during the Pleistocene.
Combinations of morphometric and molecular techniques are often used to quantify genetic diversity and to determine intra-generic boundaries of bee subpopulations (Sittipraneed et al. 2001; Arias et al. 2006; Mendes et al. 2007; Tofilski 2008; May-Itzá et al. 2012; Rattanawannee et al. 2012; Wappler et al. 2012). For instance, Francoy et al. (2006) showed that a single wing cell, the radial cell, carries sufficient information to correctly classify three groups of Apis mellifera (Africanized, Italian, and Carniolan) with a fidelity of nearly 99%. Morphometric analysis of wing venation of the Brazilian stingless bee Plebeia remota revealed cryptic species within the population (Francisco et al. 2008). Francoy et al. (2011) proposed that geometric morphometric analysis of wing shape could be used as a first step for assigning genetic lineages and geographic origins samples of the stingless bee Melipona beecheii.
Here we use a phylogenetic analysis of the COI region of the mitochondria in combination with geometric morphometrics of wing venation to determine whether the T. collina population of Thailand shows a pattern of differentiation about the Kra ecotone that parallels that of A. cerana. If so, then this would reinforce the idea that there is a sharp biological division north and south of the ecotone that acts as a barrier to gene flow and enhances differentiation of bee populations. If there is no population subdivision in T. collina, then it may be inferred that the sharp boundary observed in A. cerana arises from a hybrid zone brought about by the reunification of the Sunderland and Indo-Chinese subpopulations of A. cerana in this area after the Pleistocene.
2 Materials and methods
2.1 Sample collection
T. collina workers were sampled from 71 colonies from 25 locations throughout Thailand. Sampling locations were grouped into seven geographical subpopulations (Table I, Figure 1). The bees were collected directly at the nest entrance tube of each colony. For morphometric analysis, at least 15 workers were collected, immediately killed with ethyl acetate, and then stored in 70% (v/v) ethanol. For molecular analysis, bees were immediately preserved in 95% (v/v) ethanol and then kept at −20°C until analysis.
Tetragonilla collina collection sites in Thailand. The numbers correspond to those in Table I.
2.2 DNA extraction, amplification, sequencing, and alignment
Genomic DNA was extracted from thoracic muscle of one worker bee per colony (34 colonies) using the DNeasy® Blood & Tissue Kit (Qiagen, Germantown, MD) following the manufacturer’s instructions. We amplified a fragment of the mitochondrial cytochrome oxidase subunit I (COI) in a 50-μL final reaction volume containing 1× PCR master mix (Catalog#K0171, Fermentas Life Science), 200 nmol of forward (LCO1490: 5′-GGTCAACAAATCATAAAGATATTGG-3′) and reverse (HCO2198: 5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) primers (Folmer et al. 1994), and at least 200 ng of DNA template. Thermal profiles consisted of an initial denaturation step of 94 °C for 5 min, followed by 35 cycles of 94 °C for 1 min, 48 °C for 1 min, and 72 °C for 150 s, with a final extension step of 72 °C for 5 min.
Amplified PCR products were purified using QIAquick® Gel Extraction Kit (Qiagen, Germantown, MD) and directly sequenced by AITbiotech Pty Ltd. (The Rutherford Science Park 1, Singapore). Partial DNA sequences were aligned and edited using MEGA6 v6.06 (Tamura et al. 2013). Sequences have been deposited in GenBank under accessions KU934111–KU934146 (Table I). To obtain outgroups for phylogenetic analysis, we sequenced the same COI fragment from three additional stingless bee species [Tetragonula pagdeni Schwarz, 1939 (GenBank ID: KU934145), Homotrigona fimbriata Smith, 1857 (GenBank ID: KU934146), and Tetrigona apicalis Smith, 1857 (GenBank ID: KU934147)], all collected in Thailand.
2.3 Molecular diversity indices analysis and phylogenetic reconstruction
Measures of genetic diversity including the average number of nucleotide differences (k), number of polymorphic sites (S), haplotype diversity (h) (Nei 1987), and nucleotide diversity (π) (Nei and Li 1979) were obtained using DnaSP v5.0 (Librado and Rozas 2009).
Maximum likelihood (ML) and Bayesian inference (BI) methods were used to reconstruct the phylogenetic relationships among COI haplotypes. The program Kakusan4 (Tanabe 2007), with maximum likelihoods calculated in TREEFINDER (Jobb et al. 2004), was used to estimate the best-fit models of nucleotide substitution as determined by the Akaike information criterion, AIC, (Akaike 1974) implemented for ML and the Bayesian information criterion, BIC, (Schwarz 1978) for BI. The ML analysis was performed using the likelihood-ratchet method in TREEFINDER (Jobb et al. 2004), with 1000 bootstrap replicates to estimate branch confidence values. Tree topologies with bootstrap values 70% or greater were regarded as being sufficiently resolved (Huelsenbeck and Hillis 1993). The BI analysis was performed with MrBayes v3.1 (Huelsenbeck and Ronquis 2001), which employs a Metropolis-coupled, Markov chain Monte Carlo (MC-MCMC) sampling approach. A four-chain MC-MCMC analysis was run twice in parallel (with default heating values) for one million generations starting with a random tree, and trees were collected every 100 generations. The log-likelihood values of the sample points were plotted against the generation time, and 25% of the generations were discarded as “burn-in” samples. The remaining trees were used to estimate consensus tree topology, bipartition posterior probability (bpp), and branch length (Huelsenbeck and Ronquis 2001). A bi-partition posterior probability of 0.95 or greater was regarded as significant support for the consensus tree (Larget and Simon 1999).
2.4 Geometric morphometrics
Ten workers were randomly selected from each of 71 colonies for dissection, giving a total of 710 bees analyzed. The right forewing of each bee was dissected and slide-mounted. Wings were photographed with a digital camera attached on a stereomicroscope (Olympus SZX16) under ×25 magnification with the same camera setting and by the same person (AR). The wing images were randomly ordered using tpsUtil v.1.49 (Rohlf 2012) before bi-dimensional coordination of landmarks. A set of 13 homologous was digitized using the tpsDig2 v.2.16 software (Rohlf 2010) by the same person (AR). Landmarks are shown in Figure 2.
All specimens were digitized twice by the same person (AR). Procrustes ANOVA (Klingenberg and McIntyre 1998) was then performed using MorphoJ v.1.05c (Klingenberg 2011) to ensure that the observed variation was attributable to biological variation and not to measurement error. Repeatability (R), the proportion of variance due to true variation among individuals in relation to the total variance, was calculated according to Arnqvist and Mårtensson (1988). To reduce the effects of measurement error, repeated measurements were averaged in MorphoJ and used in subsequent analyses.
Samples were grouped into seven prior groups according to geographical area (Table I) and into two groups based on the molecular analysis (see Sect. 3). Centroid size (CS) of the landmarks were calculated as an indicator of overall size of the wing (Zelditch et al. 2004) and used to assess whether bees of different subpopulations differed in size. We compared CS of the landmarks between two of the clades suggested by the COI analysis using an independent sample t test.
For variation of wing shape, the two-dimensional landmark data were first subjected to the Procrustes superimposition, which removes variation (i.e., scale, position, and orientation) that is not attributable to wing shape variation (Dryden and Mardia 1998). Canonical variate analysis (CVA) was then performed to examine the relative difference in wing shape among populations using MorphoJ. Mahalanobis distances and Procrustes distances between pairwise populations were determined and the significance of differences assessed by a permutation test (10,000 iterations). In order to infer phenotypic relationships of wing-shape variation among populations, a neighbor-joining tree (NJ) (Saitou and Nei 1987) based on Mahalanobis distances between the centroids of each population derived from CVA was constructed using MEGA6 v6.06.
3 Results
3.1 Molecular data analysis
We obtained DNA sequence comprising 519 base pairs (bp) from 34 individuals. The nucleotide composition showed high A+T content (average 71.48%). Multi-alignment and pair-wise sequence comparisons showed a total of 119 variable sites (89 informative), with 131 single base substitution sites comprising 93 transitions (70.99%) and 38 transversions (29.01%).
Molecular diversity indices are shown in Table II. In total, 19 different haplotypes were detected from the 34 T. collina individuals examined. The estimates of haplotype diversity (h) were high overall (0.95), ranging from 0.67 to 1.00 per subpopulation (Table II). Overall nucleotide diversity (π) was 0.073, and ranged from 0.011–0.070 per population. Populations from the east had the highest levels of nucleotide diversity, while those from the south had the lowest (Table II). When the bee samples were divided into two groups based on the two major clades suggested by the phylogenetic analysis of mitochondrial COI sequences (see below), molecular diversity indices of clade A were higher than those of clade B (Table II).
3.2 Phylogenetic analysis
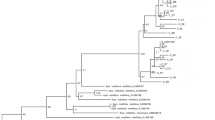
The best-fit evolutionary model for a ML tree under the AIC was J2+G+I and that for a BI tree under BIC was HKY85+G. The topology of the Bayesian tree suggests that Thai populations of T. collina are monophyletic but divided into two major clades—clade A and clade B. Clade A comprises bees from northern, central, northeastern, eastern, and western1 (PHET1 and PHET2) populations, with a ML bootstrap value of 90.7% and a Bayesian posterior probability of 0.55. Clade B includes populations from western2 (SUNGKLA1, SUNGKLA2, and RATRY), Kra, and southern populations with high support values (ML bootstrap value of 89.9% and a Bayesian posterior probability of 1.0) (Figure 3 and Table I).
3.3 Geometric morphometric analysis
Procrustes ANOVA (Klingenberg and McIntyre 1998) showed that the measurement error was low relative to overall shape variation. The between-individual mean square significantly exceeded the mean square attributable to measurement error (F 15,598 = 52.76; P < 0.0001). In addition, the repeatability of landmark acquisition was high (R = 0.96).
The bees from seven geographic populations differed significantly in wing centroid size (ANOVA: F 6, 703 = 114.8; P < 0.0001). When the bee samples were divided into two groups based on the two major clades suggested by the phylogenetic analysis of mitochondrial COI sequences (Figure 3), bees of clade A (mean wing CS = 772.64 ± 26.82) had significantly (P < 0.001 two-tailed t test) larger wings than bees from clade B (734.78 ± 21.98) (Figure 4).
The first two canonical variates explained 77.640% of total variance (Table III). Based on permutation tests for Mahalanobis and Procrustes distances, most subpopulation pairs showed significant differences in forewing shape (Table IV). In addition, individuals from clade A were generally well separated from those of clade B (Figure 5).
Scatterplot of individual scores for the first two canonical variates derived from the canonical variant analysis (CVA) of landmarks. Wireframes representing the shape change (solid line) from the consensus configuration of landmarks (dash line) to each extreme negative and positive CV scores. The symbols correspond to those in Fig. 3.
Deviations in shape from the consensus configuration along the first two CV axes of the CVA plot, as represented by wireframe graphs in Figure 5, showed that individuals located in the positive dimension of CV1 had an extended cross vein between the cubitus and vannal vein (cu-v) (landmarks 1 and 2) compared to individuals in the negative dimension. Further, the first branch of cubitus (Cu1) (landmarks 11 and 12) was constricted in the positive group relative to the negative group. Wing shape change along CV2 arose from the distal shift of landmarks 7 and 8 (radial sector) in the positive group when compared to the negative group.
A neighbor-joining tree constructed using Mahalanobis distances between population centroids revealed two distinct groups that are similar to the A and B clades revealed by the mitochondrial phylogenetic analysis (Figure 6).
4 Discussion
Both genetic and morphological data strongly suggest that the T. collina population of Thailand is divided into two distinct clades. One clade (B), which is extant on the Thai-Malay Peninsula, also extends north of the Peninsula along the Myanmar border (Figure 3). This Thai-Malay clade is significantly smaller in wing size than the second clade (A) that occupies all other areas. There is a small area of overlap between clades A and B well north of the Kra Isthmus (Figure 3).
Various studies of population genetic structure of Thai honey bees have found a biogeographical transition zone between mainland and peninsula populations focused on a sharp boundary at the Kra ecotone (at Bang Saphan, Prachuap Khiri Khan: 11° 24′ N, 99° 31′ E; and Tup Sa Kae, Prachuap Khiri Khan: 11° 31′ N, 99° 35′ E) (Limbipichai 1990; Deowanish et al. 1996; Smith and Hagen 1999; Warrit et al. 2006). Woodruff (2003) suggests that sea level rises submerged sections of the Thai-Malay Peninsula on at least two occasions during the early/middle Miocene (24–13 mya) and in the early Pliocene (5.5–4.5 mya). These historical inundations most likely led to the contemporary subdivisions in various honey bee populations, but what about stingless bees?
Thummajitsakul et al. (2008) examined genetic variation and population structure of the arboreal stingless bee, Trigona pagdeni Schwarz, based on molecular markers. They detected differentiation between samples collected north and south of the Isthmus of Kra, but a much stronger differentiation between populations in the northeast with respect to all other populations. This differentiation parallels that seen here in T. collina, and suggests that stingless bees and honey bees differ strongly in their biogeography in Thailand.
In stingless bees, gene flow via females is restricted by their reproductive biology. Stingless bee colonies propagate by establishing a new nest nearby the parent nest (Roubik 2006). Food and other resources are transferred from the parent colony to the daughter nest over several weeks or months, greatly restricting the dispersal distance (Michener 1979; Inoue et al. 1984; van Veen and Sommeijer 2000) and the extent to which mitochondrial haplotypes can spread per generation via queens (Francisco and Arias 2010; de J. May-Itzá et al. 2012; Nogueira et al. 2014). Males probably disperse further than queens (Paxton 2000; Cameron et al. 2004; Kraus et al. 2008; Mueller et al. 2012), but nonetheless the flight distance of Melipona scutellaris males, for example, is only 0.8–1 km (Carvalho-Zilse and Kerr 2004) and less than 10 km for Scaptotrigona mexicana (Kraus et al. 2008). Therefore, high population subdifferentiation is expected (Francisco et al. 2014) and usually observed (e.g., Franck et al. 2004; Quezada-Euán et al. 2007; Tavares et al. 2007; Thummajitsakul et al. 2008, 2010; de J. May-Itzá et al. 2012; Brito et al. 2014) in stingless bee populations worldwide. Geographical barriers such as rivers, oceans, and mountain ranges can further restrict the dispersal of stingless bees (Brito et al. 2014). Despite limited dispersal, the clade B of T. collina appears to have expanded its range into northwest Thailand, where it apparently out-competes the mainland clade. This may be because the forest types of western Thailand have more in common with the Malay Peninsula than with central and northern Thailand (Maxwell 2001, 2004; Wikramanayake et al. 2002).
More important than forest type may be a combination of altitude and climate. The transition zone between clades A and B appears to be the Central Plane of Thailand (Figure 3). This area is very dry in the dry season but endures prolonged periods of inundation during the wet season. As T. collina nest in cavities, either underground (typically beneath a tree), or in a termite mound (Jongjitvimol and Wattanachaiyingcharoen 2007), topology and climate combine to make the Central Plane an inhospitable environment for ground-nesting T. collina, and the area probably acts as a strong impediment to gene flow between clades A and B. We therefore speculate that this area acts as a natural barrier to gene flow between the two clades, and maintains their integrity. Thus, we propose that it is ecological factors rather than historical biogeography that drive the contemporary distribution of the clades of T. collina. In contrast, species like A. cerana that nest in tree cavities are able to survive prolonged periods of flooding. Thus, the central plane does not affect gene flow within honey bee species. Nonetheless, the structure of the T. pagdeni population of Thailand is also separated by the central plane (Thummajitsakul et al. 2008), yet this species also nests in trees above ground, and is therefore typically unaffected by seasonal flooding.
References
Akaike H. (1974) A new look at the statistical model identification. IEEE Trans. Automat. Control 19, 716–723.
Arias M.C., R.M. Brito, F.O. Francisco, G. Moretto, F.F. de Oliveira, D. Silvestre, W.S. Sheppard (2006) Molecular markers as a tool for population and evolutionary studies of stingless bees. Apidologie 37, 259–274.
Arnqvist G., T. Mårtensson (1988) Measurement error in geometric morphometrics: empirical strategies to assess and reduce its impact on measurements of shape. Acta Zool. Acad. Sci. Hung. 44, 73–96.
Brito R.M., F.O. Francisco, S.Y.W. Ho, B.P. Oldroyd (2014) Genetic architecture of the Tetragonula carbonaria species complex of Australian stingless bees (Hymenoptera: Apidae: Meliponini). Biol. J. Linn. Soc. 113, 149–161.
Cameron E.C., P. Franck, B.P. Oldroyd (2004) Genetic structure of nest aggregations and drone congregations of the south-east Asian stingless bee Trigona collina. Mol. Ecol. 13, 2357–2364.
Carvalho-Zilse G.A., W.E. Kerr (2004) Substituição natural de rainhas fisogástricas e distância de vôo dos machos em Tiuba (Melipona compressipes fasciculata Smith, 1854) e Uruçu (Melipona scutellaris Latreille, 1811) (Apidae, Meliponini). Acta Amaz. 34, 649–652.
de J. May-Itzá W., J.J.G. Quezada-Euán, R. Ayala, P. De La Rúa (2012) Morphometric and genetic analyses differentiate Mesoamerican populations of the endangered stingless bee Melipona beecheii (Hymenoptera: Meliponidae) and support their conservation as two separate units. J. Insect Conserv. 16, 723–731.
Deowanish S., J. Nakamura, M. Matsuka, K. Kimura (1996) MtDNA variation among subspecies of Apis cerana using restriction fragment length polymorphism. Apidologie 27, 407–413.
Dryden I.L., K.V. Mardia (1998) Statistical Shape Analysis. Wiley, Chichester.
Dyer F.C., T.D. Seeley (1994) Colony migration in the tropical honey bee Apis dorsata F. (Hymenoptera: Apidae). Insectes Soc. 41, 129–140.
Folmer O., M. Black, W. Hoeh, R. Lutz, R. Vrijenhoek (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Marine Biol. Biotech. 3, 294–299.
Francisco F.D., M.C. Arias (2010) Inferences of evolutionary and ecological events that influenced the population structure of Plebeia remota a stingless bee from Brazil. Apidologie 41, 216–224.
Francisco F.O., P. Nunes-Silva, T.M. Francoy, D. Wittmann, V.L. Imperatriz-Fonseca, M.C. Arias, E.D. Morgan (2008) Morphometrical, biochemical and molecular tools for assessing biodiversity. An example in Plebeia remota (Holmberg, 1903) (Apidae, Meliponini). Insectes Soc. 55, 231–237.
Francisco F.D., L.R. Santiago, R.M. Brito, B.P. Oldroyd, M.C. Arias (2014) Hybridization and asymmetrical introgression between Tetragonisca angustula and Tetragonisca fiebrigi. Apidologie 45, 1–9.
Franck P., E. Cameron, G. Good, J.-Y. Rasplus, B.P. Oldroyd (2004) Nest architecture and genetic differentiation in a species complex of Australian stingless bees. Mol. Ecol. 13, 2317–2331.
Francoy T.M., P.R.R. Prodo, L.S. Gonçalves, L.D. Costa, D. deJong (2006) Morphometric differences in a single wing cell can discriminate Apis mellifera racial types. Apidologie 37, 91–97.
Francoy T.M., M.L. Grassi, V.L. Imperatriz-Fonseca, W. de Jesús May-Itzá, J.J.G. Quezada-Euán (2011) Geometric morphometrics of the wing as a tool for assigning genetic lineages and geographic origin to Melipona beecheii (Hymenoptera: Meliponini). Apidologie 42, 499–507.
Huelsenbeck J.P., D.M. Hillis (1993) Success of phylogenetic methods in the four taxon case. Syst. Biol. 42, 247–264.
Huelsenbeck J.P., F. Ronquis (2001) MrBayes: Bayesian inference of phylogeny. Bioinformatics 17, 754–755.
Hughes J.B., P.D. Round, D.S. Woodruff (2003) The Indochinese-Sundaic faunal transition at the Isthmus of Kra: an analysis of resident forest bird species distribution. Biogeography 30, 569–580.
Inoue T., S.F. Sakagami, S. Salmah, S. Yamane (1984) The process of colony multiplication in the Sumatran stingless bee Trigona (Tetragonula) laeviceps. Biotropica 16, 100–111.
Insuan S., S. Deowanish, S. Klinbunga, S. Sittipraneed, H.A. Sylvester, S. Wongsiri (2007) Genetic differentiation of the giant honey bee (Apis dorsata) in Thailand analysed by mitochondrial genes and microsatellites. Biochem. Genet. 45, 345–361.
Itioka T., T. Inoue, H. Kaliang, M. Kato, T. Nagamitsu, K. Momose, S. Sakai, T. Yumoto, S.U. Mohamad, A.A. Hamid, S. Yamane (2001) Six-year population fluctuation of the giant honey bee Apis dorsata (Hymenoptera : Apidae) in a tropical lowland dipterocarp forest in Sarawak. Ann. Ent. Soc. Am. 94, 545–549.
Jobb G., A. von Haeseler, K. Strimmer (2004) TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol. Biol. 4, 18.
Jongjitvimol T., W. Wattanachaiyingcharoen (2007) Distribution, nesting sites and nest structures of the stingless bee species, Trigona collina Smith, 1857 (Apidae, Meliponinae) in Thailand. Nat. Hist. J. Chulalongkorn Univ. 7, 25–34.
Kerr W.E., V. Maule (1964) Geographic distribution of stingless bees and its implications. J. NY Ent. Soc. 72, 2–18.
Klingenberg C.P. (2011) MorphoJ: an integrated software package for geometric morphometrics. Mol. Ecol. Resour. 11, 353–357.
Klingenberg C.P., G.S. McIntyre (1998) Geometric morphometrics of developmental instability: analyzing patterns of fluctuating asymmetry with Procrustes methods. Evolution 52, 1363–1375.
Koeniger N., G. Koeniger (1980) Observations and experiments on migration and dance communication of Apis dorsata in Sri Lanka. J. Apic. Res. 19, 21–34.
Kraus F.B., S. Weinhold, R.F.A. Moritz (2008) Genetic structure of drone congregations of the stingless bee Scaptotrigona mexicana. Insectes Soc. 55, 22–27.
Larget B., D.L. Simon (1999) Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Mol. Biol. Evol. 16, 750–759.
Librado P., J. Rozas (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452.
Limbipichai K., 1990. Morphometric studies on the eastern honey bee (Apis cerana Fabricius) in Thailand and the Malaysian peninsula. Masters Thesis, Department of Biology, Chulalongkorn University, Bangkok.
Maxwell J.F. (2001) A reassessment of the forest types of Thailand. In: Maxwell J.F.. Elliot S. (Eds.), Vegetation and vascular flora of Doi Sutep-Pui National Park, northern Thailand. Biodiversity Research and Training Program Bangkok, pp. 1–17
Maxwell J.F. (2004) A synopsis of the vegetation of Thailand. Nat. Hist. J. Chulalongkorn Univ. 4, 19–29.
May-Itzá W.D.J., J.J.G. Quezada-Euan, R. Ayala, P. De la Rúa (2012) Morphometric and genetic analyses differentiate Mesoamerican populations of the endangered stingless bee Melipona beecheii (Hymenoptera: Meliponidae) and support their conservation as two separate units. J. Insect Conserv. 16, 723–731.
Mendes M.F.M., T.M. Francoy, P. Nunes-Silva, C. Menezes, V.L. Imperatriz-Fonseca (2007) Intra-populational variability in Nannotrigona testaceicornis Lepeletier, 1836 (Hymenoptera, Meliponini) using relative warps analysis. Bioscience J. 23, 147–152.
Michener C.D. (1979) Biogeography of the bees. Ann. Missouri Bot. Gard. 66, 277–347.
Mueller M.Y., R.F.A. Moritz, F.B. Kraus (2012) Outbreeding and lack of temporal genetic structure in a drone congregation of the neotropical stingless bee Scaptotrigona mexicana. Ecol. Evol. 2, 1304–1311.
Nakamura J., S. Wongsiri, M. Sasaki (1991) Apis cerana on Samui Island and its beekeeping. Honeybee Science 12, 27–30.
Nei M. (1987) Molecular Evolutionary Genetics. Columbia University Press, New York.
Nei M., W.H. Li (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Nat. Acad. Sci. USA 76, 5269–5273.
Nogueira J., J.C. Ramos, J. Benevenuto, T.M. Fernandes-Salomão, H.C. Resende, L.A.O. Campos, M.G. Tavares (2014) Conservation study of an endangered stingless bee (Melipona capixaba—Hymenoptera: Apidae) with restricted distribution in Brazil. J. Insect Conserv. 18, 317–326.
Paar J., B.P. Oldroyd, E. Huettinger, G. Kastberger (2004) Genetic structure of an Apis dorsata population: the significance of migration and colony aggregation. J. Hered. 95, 119–126.
Paxton R.J. (2000) Genetic structure of colonies and male aggregation in the stingless bee Scaptotrigona postica, as revealed by microsatellite analysis. Insectes Soc. 47, 63–69.
Quezada-Euán J.J.G., R.J. Paxton, K.A. Palmer, W.D.J.M. Itzá, W.T. Tay, B.P. Oldroyd (2007) Morphological and molecular characters reveal differentiation in a Neotropical social bee, Melipona beecheii (Apidae: Meliponini). Apidologie 38, 247–258.
Rattanawannee A., C. Chanchao, S. Wongsiri (2012) Geometric morphometric analysis of giant honeybee (Apis dorsata Fabricius, 1793) populations in Thailand. J. Asia Pac. Entomol. 15, 611–618.
Roberts T.E. (2011) Molecular phylogeny of treeshrews (Mammalia: Scandentia) and the timescale of diversification in Southeast Asia. Mol. Phylo. Evol. 60, 358–372.
Rohlf F.J., 2010. TpsDig, Department of Ecology and Evolution, State University of New York at Stony Brook, New York.
Rohlf F.J., 2012. tpsUtil, Department of Ecology and Evolution, State University of New York at Stony Brook, New York.
Roubik D.W. (2006) Stingless bee nesting biology. Apidologie 37, 124–143.
Saitou N., M. Nei (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sakagami S., S.G. Khoo (1987) Taxonomic status of the Malesian stingless bee Trigona reepeni, with discovery of Trigona pagdeni from northern Malaya. Jap. J. Entomol. (Kontyû) 55, 207–214.
Schwarz G.E. (1978) Estimating the dimension of a model. Ann. Stat. 6, 461–464.
Sihanuntavong D., S. Sittipraneed, S. Klinbunga (1999) Mitochondrial DNA diversity and population structure of the honey bee, Apis cerana, in Thailand. J. Apic. Res. 38, 211–219.
Sittipraneed S., S. Laoaroon, S. Klinbunga, S. Wongsiri (2001) Genetic differentiation of the honey bee (Apis cerana) in Thailand: evidence from microsatellite polymorphism. J. Apic. Res. 40, 9–16.
Smith D.R., R.H. Hagen (1996) The biogeography of Apis cerana as revealed by mitochondrial DNA sequence data. J. Kans. Ent. Soc. 69, 294–310.
Smith D.R., R.H. Hagen (1999) Phylogeny and biogeography of Apis cerana subspecies: testing alternative hypotheses, in: Hoopingarner R., Connor L. (Eds.), Apiculture for the 21st Century, Wicwas Press, Cheshire, pp. 60–68.
Tamura K., G. Stecher, D. Peterson, A. Filipski, S. Kumar (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729.
Tanabe A.S. (2007) Kakusan: a computer program to automate the selection of a nucleotide substitution model and the configuration of a mixed model on multilocus data. Mol. Ecol. Notes 7, 962–964.
Tavares M.G., L.A. Santos, B.A. Arantes, L.D. Meneses, B.A.H. Pereira, C.R. Guimarães, T.M. Fernandes-Salomão, L.A. de Oliveira-Campos (2007) Genetic divergence between populations of the stingless bee uruçu amarela (Melipona rufiventris group, Hymenoptera, Meliponini): Is there a new Melipona species in the Brazilian state of Minas Gerais? Genet. Mol. Biol. 30, 667–675.
Theeraapisakkun M., S. Klinbunga, S. Sittipraneed (2010) Development of a species-diagnostic marker and its application for population genetics studies of the stingless bee Trigona collina in Thailand. Genet Mol Res 9, 913–930.
Thummajitsakul S., S. S. Klinbunga, D. Smith, S. Sittipraneed (2008) Genetic diversity and population structure of Trigona pagdeni Schwarz in Thailand. Apidologie 39, 446–455.
Thummajitsakul S., S. Klinbunga, S. Sittipraneed (2010) Development of a species-diagnostic marker for identification of the stingless bee Trigona pagdeni in Thailand. Biochem. Genet. 48, 181–192.
Tofilski A. (2008) Using geometric morphometrics and standard morphometry to discriminate three honeybee subspecies. Apidologie 39, 558–563.
van Veen J.W., M.J. Sommeijer (2000) Colony reproduction in Tetragonisca angustula (Apidae, Meliponini). Insectes Soc. 47, 70–75.
Wappler T., T.D. Meulemeester, A.M. Aytekin, D. Michez, M.S. Engel (2012) Geometric morphometric analysis of a new Miocene bumble bee from the Randeck Maar of southwestern Germany (Hymenoptera: Apidae). Syst. Entomol. 37, 784–792.
Warrit N., D.R. Smith, C. Lekprayoon (2006) Genetic subpopulations of Varroa mites and their Apis cerana hosts in Thailand. Apidologie 37, 19–30.
Wikramanayake E., E. Dinerstein, C.L. Loucks (2002) Terrestrial Ecoregions of the Indo-Pacific. Island Press, Washington D.C.
Woodruff D.S. (2003) Neogene marine transgressions, paleogeography and biogeographic transition on the Thai-Malay Peninsula. J. Biogeogr. 30, 551–567.
Zelditch M.L., D.L. Swiderski, H.D. Sheets, W.L. Fink (2004) Geometric Morphometrics for Biologists: a primer. Elsevier Academic Presss, San Diego.
Acknowledgements
We thank the Department of Entomology, Faculty of Agriculture, Kasetsart University (Bang Khan campus), and The Center of Excellence in Biodiversity, Chulalongkorn University, for providing facilities and laboratory support. We also acknowledge the assistance of Sarawoot Gomuttapong, who recently passed away, for his help with field collections. This study was supported by grant from the Thailand Research Fund (grant no. MRG6080272) and National Research University Project (NRU) of the Thailand’s Office of the Higher Education Commission
Author information
Authors and Affiliations
Contributions
AR and OD conceived the study and collected the data. AR and EJ analyzed the data. AR and BPO wrote the paper. All authors approved the final manuscript.
Corresponding author
Additional information
Manuscript editor: Marina Meixner
Le séquençage mitochondrial et la morphométrie géométrique suggèrent l’existence de 2 taxons monophylétiques dans la population de Tetragonilla collina (Apidae: Meloponini) de Thaïlande
Abeille sans aiguillon / variation génétique / biogéographie / Isthme de Kra
Mitochondriale Sequenzdaten und geometrische Morphometrie deuten auf zwei monophyletische Gruppen in der Population von Tetragonilla collina (Apidae: Meliponini) in Thailand hin
Stachellose Bienen / genetische Variation / geometrische Morphometrie / Biogeographie / Isthmus von Kra
Rights and permissions
About this article
Cite this article
Rattanawannee, A., Jeratthitikul, E., Duangpakdee, O. et al. Mitochondrial sequencing and geometric morphometrics suggest two clades in the Tetragonilla collina (Apidae: Meliponini) population of Thailand. Apidologie 48, 719–731 (2017). https://doi.org/10.1007/s13592-017-0517-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13592-017-0517-3