Abstract
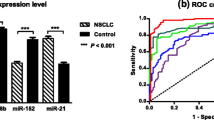
Global deregulation in miRNA expression is a hallmark of cancer cell. An estimated 2300 mature miRNAs are encoded by human genome; role of many of which in carcinogenesis and as cancer biomarkers remains unexplored. In this study, we investigated the utility of miR-3692-3p, miR-3195, and miR-1249-3p as biomarkers in non-small cell lung cancer (NSCLC). For this prospective study, 115 subjects, including 75 NSCLC patients and 40 controls, were recruited. The expression of miR-3692-3p, miR-3195, and miR-1249-3p was checked using qRT-PCR. The miRNA expression was correlated with survival outcome and therapeutic response. There were no significant differences in the mean age of NSCLC patients and controls (56.2 and 55.3 years, respectively; p = 0.3242). Majority of NSCLC patients (67%) were smokers. We observed a significant upregulation of miR-3692-3p expression (p < 0.0001), while the expression of miR-3195 (p = 0.0017) and miR-1249-3p was significantly downregulated (p < 0.0001) in the serum of NSCLC patients as compared to controls. The expression of miR-1249-3p was significantly upregulated in lung adenocarcinoma versus lung squamous cell carcinoma (p = 0.0178). Interestingly, patients who responded to chemotherapy had higher expression of miR-1249-3p than non-responders (p = 0.0107). Moreover, patients with higher expression of miR-3195 had significantly longer overall survival (p = 0.0298). In multivariate analysis, miR-3195 emerged as independent prognostic factor for overall survival. We conclude that the miR-3195 may have prognostic significance, while miR-1249-3p may predict therapeutic response in NSCLC. Further studies are warranted to elucidate the role of these miRNAs in lung carcinogenesis and their utility as candidate cancer biomarkers.


Similar content being viewed by others
Abbreviations
- NSCLC:
-
Non-small cell lung cancer
- LUSC:
-
Lung squamous cell carcinoma
- LUAD:
-
Lung adenocarcinoma
- miRNA:
-
MicroRNA
- RT:
-
Room temperature
- qRT-PCR:
-
Reverse transcription quantitative polymerase chain reaction
- OS:
-
Overall survival
- PFS:
-
Progression-free survival
- ROC:
-
Receiver operating characteristic
- AUC:
-
Area under the curve
- ECOG PS:
-
Eastern Cooperative Oncology Group performance status
- BC:
-
Breast cancer
- HCC:
-
Hepatocellular cancer
- PC:
-
Prostate cancer
- GC:
-
Gastric cancer
- BCR:
-
Biochemical recurrence
- ICOS:
-
Inducible T-cell co-stimulator
- UTR:
-
Untranslated region
- CRC:
-
Colorectal cancer
- HBV:
-
Hepatitis virus B
- SP:
-
Side population
- HOXB8:
-
Homeobox 8
- VEGFA:
-
Vascular endothelial growth factor A
- HMGA2:
-
High-mobility group AT-hook 2
- APC2:
-
Adenomatous polyposis coli 2
- Gli1:
-
Glioma-associated oncogene 1
- PTCH-1:
-
Patched-1
References
Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. https://doi.org/10.3322/caac.21492.
Ramalingam SS, Owonikoko TK, Khuri FR. Lung cancer: new biological insights and recent therapeutic advances. CA Cancer J Clin. 2011;61:91–112. https://doi.org/10.3322/caac.20102.
Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–80. https://doi.org/10.1016/S1470-2045(10)70087-5.
Korpanty GJ, Graham DM, Vincent MD, et al. Biomarkers that currently affect clinical practices in lung cancer: EGFR, ALK, MET, ROS-1, and KRAS. Front Oncol. 2014;4:204. https://doi.org/10.3389/fonc.2014.00204.
Brahmer JR, Govindan R, Anders RA, et al. The Society for Immunotherapy of Cancer consensus statement on immunotherapy for the treatment of non-small cell lung cancer (NSCLC). J Immunother Cancer. 2018;6(1):75. https://doi.org/10.1186/s40425-018-0382-2.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. https://doi.org/10.1016/s0092-8674(04)00045-5.
Kasinski AL, Slack FJ. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–64. https://doi.org/10.1038/nrc3166.
Singh DK, Bose S, Kumar S. Role of microRNA in regulating cell signaling pathways, cell cycle, and apoptosis in non-small cell lung cancer. Curr Mol Med. 2016;16:474–86. https://doi.org/10.2174/1566524016666160429120702.
Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–59. https://doi.org/10.1002/emmm.201100209.
Rupaimoole R, Calin GA, Lopez-Berestein G, et al. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6:235–46. https://doi.org/10.1158/2159-8290.CD-15-0893.
He Y, Lin J, Kong D, et al. Current state of circulating microRNAs as cancer biomarkers. Clin Chem. 2015;61:1138–55. https://doi.org/10.1373/clinchem.2015.241190.
Alles J, Fehlmann T, Fischer U, et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019;47(7):3353–64. https://doi.org/10.1093/nar/gkz097.
Kumar S, Sharawat SK, Ali A, et al. Identification of differentially expressed circulating serum microRNA for the diagnosis and prognosis of Indian non-small cell lung cancer patients. Curr Probl Cancer. 2020. https://doi.org/10.1016/j.currproblcancer.2020.100540.
Detterbeck FC, Boffa DJ, Kim AW, et al. The eighth edition lung cancer stage classification. Chest. 2017;151:193–203. https://doi.org/10.1016/j.chest.2016.10.010.
Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228–47. https://doi.org/10.1016/j.ejca.2008.10.026.
Gao P, Teng Z, Ji L, et al. Interactions of ABLIMI and CXCL5 with miRNAs as a prognostic indicator for clinical outcome of osteosarcoma. Int J Clin Exp Med. 2016;9(8):15345–53.
He Z, Yi J, Liu X, et al. MiR-143-3p functions as a tumor suppressor by regulating cell proliferation, invasion and epithelial-mesenchymal transition by targeting QKI-5 in esophageal squamous cell carcinoma. Mol Cancer. 2016;15(1):51. https://doi.org/10.1186/s12943-016-0533-3.
Kim BG, Kang S, Han HH, et al. Transcriptome-wide analysis of compression-induced microRNA expression alteration in breast cancer for mining therapeutic targets. Oncotarget. 2016;7(19):27468–78. https://doi.org/10.18632/oncotarget.8322.
Stückrath I, Rack B, Janni W, et al. Aberrant plasma levels of circulating miR-16, miR-107, miR-130a and miR-146a are associated with lymph node metastasis and receptor status of breast cancer patients. Oncotarget. 2015;6(15):13387–40101. https://doi.org/10.18632/oncotarget.3874.
Yasui T, Yanagida T, Ito S, et al. Unveiling massive numbers of cancer-related urinary-microRNA candidates via nanowires. Sci Adv. 2017;3(12):e1701133. https://doi.org/10.1126/sciadv.1701133.
Liu S, Pan H, Cao J, et al. MicroRNA-134 inhibits HCC cell growth and migration through the AKT/GSK3β/SNAIL signaling pathway. Int J Clin Exp Pathol. 2016;9(7):6877–86.
Stuopelytė K, Daniūnaitė K, Jankevičius F, et al. Detection of miRNAs in urine of prostate cancer patients. Medicina (Kaunas). 2016;52(2):116–24. https://doi.org/10.1016/j.medici.2016.02.007.
Wu D, Tang R, Qi Q, et al. Five functional polymorphisms of B7/CD28 co-signaling molecules alter susceptibility to colorectal cancer. Cell Immunol. 2015;293(1):41–8. https://doi.org/10.1016/j.cellimm.2014.11.006.
Marinelli O, Nabissi M, Morelli MB, et al. ICOS-L as a potential therapeutic target for cancer immunotherapy. Curr Protein Pept Sci. 2018;19(11):1107–13. https://doi.org/10.2174/1389203719666180608093913.
Soldevilla MM, Villanueva H, Meraviglia-Crivelli D, et al. ICOS costimulation at the tumor site in combination with CTLA-4 blockade therapy elicits strong tumor immunity. Mol Ther. 2019;27(11):1878–91. https://doi.org/10.1016/j.ymthe.2019.07.013.
Pan HL, Wen ZS, Huang YC, et al. Down-regulation of microRNA-144 in air pollution-related lung cancer. Sci Rep. 2015;5:14331. https://doi.org/10.1038/srep14331.
Huang L, Cai JL, Huang PZ, et al. miR19b-3p promotes the growth and metastasis of colorectal cancer via directly targeting ITGB8. Am J Cancer Res. 2017;7(10):1996–2008.
Schneider A, Victoria B, Lopez YN, et al. Tissue and serum microRNA profile of oral squamous cell carcinoma patients. Sci Rep. 2018;8(1):675. https://doi.org/10.1038/s41598-017-18945-z.
Morishita A, Iwama H, Fujihara S, et al. MicroRNA profiles in various hepatocellular carcinoma cell lines. Oncol Lett. 2016;12(3):1687–92. https://doi.org/10.3892/ol.2016.4853.
Hide T, Komohara Y, Miyasato Y, et al. Oligodendrocyte progenitor cells and macrophages/microglia produce glioma stem cell niches at the tumor border. EBioMedicine. 2018;30:94–104. https://doi.org/10.1016/j.ebiom.2018.02.024.
Sohn EJ, Won G, Lee J, et al. Upregulation of miRNA3195 and miRNA374b mediates the anti-angiogenic properties of melatonin in hypoxic PC-3 prostate cancer cells. J Cancer. 2015;6:19–28. https://doi.org/10.7150/jca.9591.
Zhu C, Huang Q, Zhu H. Melatonin inhibits the proliferation of gastric cancer cells through regulating the miR-16-5p-Smad3 pathway. DNA Cell Biol. 2018;37(3):244–52. https://doi.org/10.1089/dna.2017.4040.
Jiang J, Ma B, Li X, et al. MiR-1281, a p53-responsive microRNA, impairs the survival of human osteosarcoma cells upon ER stress via targeting USP39. Am J Cancer Res. 2018;8(9):1764–74.
Liu J, Ma L, Wang Z, et al. MicroRNA expression profile of gastric cancer stem cells in the MKN-45 cancer cell line. Acta Biochim Biophys Sin (Shanghai). 2014;46(2):92–9. https://doi.org/10.1093/abbs/gmt135.
Chen Y, Zhao J, Luo Y, et al. Downregulated expression of miRNA-149 promotes apoptosis in side population cells sorted from the TSU prostate cancer cell line. Oncol Rep. 2016;36(5):2587–600. https://doi.org/10.3892/or.2016.5047.
Qin X, Yu S, Xu X, et al. Comparative analysis of microRNA expression profiles between A549, A549/DDP and their respective exosomes. Oncotarget. 2017;8(26):42125–35. https://doi.org/10.18632/oncotarget.15009.
Wozniak M, Sztiller-Sikorska M, Czyz M. Expression of miRNAs as important element of melanoma cell plasticity in response to microenvironmental stimuli. Anticancer Res. 2015;35(5):2747–58.
Ghosh T, Varshney A, Kumar P, et al. MicroRNA-874-mediated inhibition of the major G1/S phase cyclin, CCNE1, is lost in osteosarcomas. J Biol Chem. 2017;292(52):21264–81. https://doi.org/10.1074/jbc.M117.808287.
Dankert JT, Wiesehöfer M, Czyrnik ED, et al. The deregulation of miR-17/CCND1 axis during neuroendocrine transdifferentiation of LNCaP prostate cancer cells. PLoS ONE. 2018;13(7):e0200472. https://doi.org/10.1371/journal.pone.0200472.
Ding J, Wu W, Yang J, et al. Long non-coding RNA MIF-AS1 promotes breast cancer cell proliferation, migration and EMT process through regulating miR-1249-3p/HOXB8 axis. Pathol Res Pract. 2019;215(7):152376. https://doi.org/10.1016/j.prp.2019.03.005.
Chen X, Zeng K, Xu M, et al. P53-induced miR-1249 inhibits tumor growth, metastasis, and angiogenesis by targeting VEGFA and HMGA2. Cell Death Dis. 2019;10(2):131. https://doi.org/10.1038/s41419-018-1188-3.
Katayama Y, Maeda M, Miyaguchi K, et al. Identification of pathogenesis-related microRNAs in hepatocellular carcinoma by expression profiling. Oncol Lett. 2012;4(4):817–23. https://doi.org/10.3892/ol.2012.810.
Fang B, Li G, Xu C, et al. MicroRNA miR-1249 downregulates adenomatous polyposis coli 2 expression and promotes glioma cells proliferation. Am J Transl Res. 2018;10(5):1324–36.
Shu H, Hu J, Deng H. miR-1249-3p accelerates the malignancy phenotype of hepatocellular carcinoma by directly targeting HNRNPK. Mol Genet Genom Med. 2019;7(10):e00867. https://doi.org/10.1002/mgg3.867.
Seshachalam VP, Sekar K, Hui KM. Insights into the etiology-associated gene regulatory networks in hepatocellular carcinoma from The Cancer Genome Atlas. J Gastroenterol Hepatol. 2018;33(12):2037–47. https://doi.org/10.1111/jgh.14262.
Ye Y, Wei Y, Xu Y, et al. Induced MiR-1249 expression by aberrant activation of Hedegehog signaling pathway in hepatocellular carcinoma. Exp Cell Res. 2017;355(1):9–17. https://doi.org/10.1016/j.yexcr.2017.03.010.
Chen X, Xiong W, Li H. Comparison of microRNA expression profiles in K562-cells-derived microvesicles and parental cells, and analysis of their roles in leukemia. Oncol Lett. 2016;12(6):4937–48. https://doi.org/10.3892/ol.2016.5308.
Yoshii S, Hayashi Y, Iijima H, et al. Exosomal microRNAs derived from colon cancer cells promote tumor progression by suppressing fibroblast TP53 expression. Cancer Sci. 2019;110(8):2396–407. https://doi.org/10.1111/cas.14084.
Okumura T, Shimada Y, Omura T, et al. MicroRNA profiles to predict postoperative prognosis in patients with small cell carcinoma of the esophagus. Anticancer Res. 2015;35(2):719–27.
Scaravilli M, Porkka KP, Brofeldt A, et al. MiR-1247-5p is overexpressed in castration resistant prostate cancer and targets MYCBP2. Prostate. 2015;75(8):798–805. https://doi.org/10.1002/pros.22961.
Acknowledgements
The authors gratefully acknowledge the financial support from the Science and Engineering Research Board (SERB), Govt. of India, New Delhi (Grant no. SB/YS/LS-348/2013) and All India Institute of Medical Sciences, New Delhi (Grant no. A-516).
Funding
The authors gratefully acknowledge the financial support from the Science & Engineering Research Board (SERB), Govt. of India, New Delhi (Grant no. SB/YS/LS-348/2013) and All India Institute of Medical Sciences, New Delhi (Grant no. A-516).
Author information
Authors and Affiliations
Contributions
SK, VG, and MP performed all the laboratory work. SK and SKS analyzed and interpreted the experimental data. AS, PSM, SK, AM and RG collected, analyzed, and interpreted the clinical data. SK designed the study and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The study was approved by ethics committee of All India Institute of Medical Sciences, New Delhi (Ref. No. IEC-155/07.04.2017, RP-13/2017). This article does not contain any studies with animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kumar, S., Sharawat, S.K., Ali, A. et al. Differential expression of circulating serum miR-1249-3p, miR-3195, and miR-3692-3p in non-small cell lung cancer. Human Cell 33, 839–849 (2020). https://doi.org/10.1007/s13577-020-00351-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-020-00351-9