Abstract
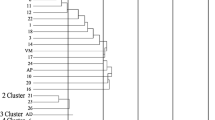
Groundnut is a rich source of several nutritional components including polyphenols and antioxidants that offer various health benefits. In this regard, the mini core accessions along with elite varieties of groundnut were used to assess genetic diversity using AhTE markers. The phenotypic observation on eight morphological, six productivity and two nutraceutical traits [total polyphenol content (TPC) and total antioxidant activity (AOA)] were studied. Correlation studies revealed a significant positive correlation between TPC and AOA. The degree of divergence with respect to nutraceutical content among the genotypes of mini core collection and elite cultivars is evident from the current study. The STRUCTURE analyses revealed the grouping of genotypes into three distinct clusters mainly based on the botanical types of groundnut. The analysis of molecular variance displayed maximum variation (97%) within the individuals of subpopulations and minimum variation (3%) among subpopulations. Principal component analysis exhibited 3 principal components that accounted for 42.17% of the total variation. Association mapping study indicated 20 significant marker-trait associations at 1% probability. The study has also identified significant marker-trait associations with nutraceutical properties of groundnut, AhTE0465-TPC and AhTE0381- AOA with explained phenotypic variation of 7.45% and 6.85% respectively. These markers were found to have positions at A02 and A09 with bHLH DNA-binding family protein and chitinase putative functions respectively. The markers associated with TPC and AOA can further be utilized for genomics-assisted breeding for nutritionally rich cultivars in groundnut.



Similar content being viewed by others
Abbreviations
- TPC:
-
Total polyphenol content
- AOA:
-
Total antioxidant activity
- AhTE:
-
Arachis hypogaea Transposable elements
- ICRISAT:
-
International crops research institute for the semi-arid tropics
- PIC:
-
Polymorphic information content
- GA:
-
Gentic advance
- GAM:
-
Genetic advance as a percent of mean
- PCA:
-
Principal component analysis
- PCoA:
-
Principal coordinate analysis
- AMOVA:
-
Analysis of molecular variance
- MTAs:
-
Marker-trait associations
- QTL:
-
Quantitative trait loci
References
Adhikari B, Dhungana SK, Ali MW, Adhikari A, Kim I, Shin D (2018) Resveratrol, total phenolic and flavonoid contents, and antioxidant potential of seeds and sprouts of Korean peanuts. Food Sci Biotechnol 27(5):1275–1284
Adhikari B, Dhungana SK, Ali MW, Adhikari A, Kim I, Shin D (2019) Antioxidant activities, polyphenol, flavonoid, and amino acid contents in peanut shell. J Saudi Soc Agric Sci 18:437–442
Arya SS, Salve AR, Chauhan S (2016) Peanuts as functional food: a review. J Food Sci Technol 53(1):31–41
Bilal M, Xiaobo Z, Arslan M, Tahir HE, Sun Y, Aadil RM (2021) Near infrared spectroscopy coupled chemometric algorithms for prediction of the antioxidant activity of peanut seed (Arachis hypogaea). J near Infrared Spectrosc 29(4):191–200
Bodoira R, Cittadini MC, Velez A, Rossi Y, Montenegro M, Martínez M, Maestri D (2022) An overview on extraction, composition, bioactivity and food applications of peanut phenolics. Food Chem 381:132250
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23(19):2633–2635
Chu Y, Chee P, Isleib TG, Holbrook CC, Ozias-Akins P (2020) Major seed size QTL on chromosome A05 of peanut (Arachis hypogaea) is conserved in the US mini core germplasm collection. Mol Breed 40(1):1–16
Chukwumah Y, Walker LT, Verghese M (2009) Peanut skin color: a biomarker for total polyphenolic content and antioxidative capacities of peanut cultivars. Int J Mol Sci 10(11):4941–4952
Dean LL, Davis JP, Shofran BG, Sanders TH (2008) Phenolic profiles and antioxidant activity of extracts from peanut plant parts. Nat Prod J 1(1):1–6
Earl DA, Vonholdt BM (2012) Structure harvester: a website and program for visualizing Structure output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8):2611–2620
Gangurde SS, Pasupuleti J, Parmar S, Variath MT, Bomireddy D, Manohar SS, Varshney RK, Singam P, Guo B, Pandey MK (2023) Genetic mapping identifies genomic regions and candidate genes for seed weight and shelling percentage in groundnut. Front Genet 14:1128182
Gomes RLF, de Almeida Lopes ÂC (2005) Correlations and path analysis in peanut. Crop Breed Appl Biotechnol 5:105–112
Gonzalez CA, Salas-Salvado J (2006) The potential of nuts in the prevention of cancer. Br J Nutr 96:S87–S94
Hake AA, Shirasawa K, Yadawad A, Sukruth M, Patil M, Nayak SN, Lingaraju S, Patil PV, Nadaf HL, Gowda MVC, Bhat RS (2017) Mapping of important taxonomic and productivity traits using genic and non-genic transposable element markers in peanut (Arachis hypogaea L.). PLoS ONE 12(10):e0186113
Hildebrand CE, Torney DC, Wagner RP (1992) Informativeness of polymorphic DNA markers. Los Alamos Sci 20:100–102
Jones JB, Provost M, Keaver L, Breen C, Ludy MJ, Mattes RD (2014) A randomized trial on the effects of flavorings on the health benefits of daily peanut consumption. Am J Clin Nutr 99(3):490–496
Khera P, Upadhyaya HD, Pandey MK, Roorkiwal M, Sriswathi M, Janila P, Guo Y, McKain MR, Nagy ED, Knapp SJ (2013) Single nucleotide polymorphism–based genetic diversity in the reference set of peanut (Arachis spp) by developing and applying cost-effective kompetitive allele specific polymerase chain reaction genotyping assays. Plant Genome. https://doi.org/10.3835/plantgenome2013.06.0019
Kolekar RM, Sujay V, Shirasawa K, Sukruth M, Khedikar YP, Gowda MVC, Pandey MK, Varshney RK, Bhat RS (2016) QTL mapping for late leaf spot and rust resistance using an improved genetic map and extensive phenotypic data on a recombinant inbred line population in peanut (Arachis hypogaea L.). Euphytica 209(1):147–156
Limmongkon A, Janhom P, Amthong A, Kawpanuk M, Nopprang P, Poohadsuan J, Somboon T, Saijeen S, Surangkul D, Srikummool M, Boonsong T (2017) Antioxidant activity, total phenolic, and resveratrol content in five cultivars of peanut sprouts. Asian Pac J Trop Biomed 7(4):332–338
Liu N, Guo J, Zhou X, Wu B, Huang L, Luo H, Chen Y, Chen W, Lei Y, Huang Y, Liao B (2020) High-resolution mapping of a major and consensus quantitative trait locus for oil content to a~ 0.8-Mb region on chromosome A08 in peanut (Arachis hypogaea L.). Theor Appl Genet 133(1):37–49
Mace ES, Bhuariwalla KK, Bhuariwalla HK, Crouch JH (2003) A high-throughput DNA extraction protocol for tropical molecular breeding programs. Plant Mol Biol Rep 21(4):459–460
Mitra M, Gantait S, Kundu R (2021) Genetic variability, character association and genetic divergence in groundnut (Arachis hypogaea L.) accessions. Legum Res 44(2):164–169
Mondal S, Badigannavar AM (2018) Mapping of a dominant rust resistance gene revealed two R genes around the major rust QTL in cultivated peanut (Arachis hypogaea L.). Theor Appl Genet 131(8):1671–1681
Mondal S, Badigannavar AM, D’Souza SF (2012) Molecular tagging of a rust resistance gene in cultivated groundnut (Arachis hypogaea L.) introgressed from Arachis cardenasii. Mol Breed 29(2):467–476
Mondal S, Phadke RR, Badigannavar AM (2015) Genetic variability for total phenolics, flavonoids and antioxidant activity of testaless seeds of a peanut recombinant inbred line population and identification of their controlling QTLs. Euphytica 204(2):311–321
Moreno JP, Johnston CA, El-Mubasher AA, Papaioannou MA, Tyler C, Gee M, Foreyt JP (2013) Peanut consumption in adolescents is associated with improved weight status. Nutr Res 33(7):552–526
Morris GP, Ramu P, Deshpande SP, Hash CT, Shah T, Upadhyaya HD, Riera-Lizarazu O, Brown PJ, Acharya CB, Mitchell SE, Harriman J (2013) Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc Natl Acad Sci 110:453–458
Mukri G, Nadaf HL, Gowda MVC, Bhat RS, Upadhyaya HD, Sujay V (2012) Phenotypic and molecular dissection of ICRISAT mini core collection of peanut (Arachis hypogaea L.) for high oleic acid. Plant Breed 131:418–422
Nabi RBS, Cho KS, Tayade R, Oh KW, Lee MH, Kim JI, Kim S, Pae SB, Oh E (2021) Genetic diversity analysis of Korean peanut germplasm using 48K SNPs ‘Axiom_Arachis’ array and its application for cultivar differentiation. Sci Rep 11(1):16630
Nayak SN, Hebbal V, Bharati P, Nadaf HL, Naidu GK, Bhat RS (2020) Profiling of nutraceuticals and proximates in peanut genotypes differing for seed coat color and seed size. Front Nutr 7:45
Nayak SN, Aravind B, Malavalli SS, Sukanth BS, Poornima R, Bharati P, Hefferon K, Kole C, Puppala N (2021) Omics technologies to enhance plant based functional foods: an overview. Front Genet 12:742095
Pandey MK, Agarwal G, Kale SM, Clevenger J, Nayak SN, Sriswathi M, Chitikineni A, Chavarro C, Chen X, Upadhyaya HD, Vishwakarma MK, Leal-Bertioli S et al (2017) Development and evaluation of a high density genotyping axiom Arachis array with 58 K SNPs for accelerating genetics and breeding in groundnut. Sci Rep 7:40577
Pandey MK, Gangurde SS, Sharma V, Pattanashetti SK, Naidu GK, Faye I, Hamidou F, Desmae H, Kane NA, Yuan M, Vadez V (2021) Improved genetic map identified major QTLs for drought tolerance-and iron deficiency tolerance-related traits in groundnut. Genes 12(1):37
Pattanashetti SK, Naidu GK, Prakyath Kumar KV, Singh OK, Biradar BD (2018) Identification of iron deficiency chlorosis tolerant sources from mini-core collection of groundnut (Arachis hypogaea L.). Plant Genet Resour 16(5):446–458
Peakall ROD, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6(1):288–295
Perrier X, Jacquemoud-Collet JP (2006) DARwin software. www.darwin.cirad.fr/darwin
Pritchard JK, Stephens M, Rosenberg NA, Donnelly P (2000) Association mapping in structured populations. Am J Hum Genet 67(1):170–181
RStudio Team (2020) RStudio: Integrated Development for R. RStudio, PBC, Boston, MA URL http://www.rstudio.com/.
Saleem MA, Naidu GK, Nadaf HL, Tippannavar PS (2018) Assessment of groundnut (Arachis hypogaea L.) mini core for resistance to multiple biotic stresses under hot spot location. Int J Curr Microbiol Appl Sci 7:1599–1614
Saleem MA, Naidu GK, Nadaf HL, Tippannavar PS (2019) Diverse sources of resistance to Spodoptera litura (F.) in groundnut. Indian J Genet 79(1):192–196
Shirasawa K, Hirakawa H, Tabata S, Hasegawa M, Kiyoshima H, Suzuki S, Sasamoto S, Watanabe A, Fujishiro T, Isobe S (2012a) Characterization of active miniature inverted-repeat transposable elements in the peanut genome. Theor Appl Genet 124(8):1429–1438
Shirasawa K, Koilkonda P, Aoki K, Hirakawa H, Tabata S, Watanabe M, Hasegawa M, Kiyoshima H, Suzuki S, Kuwata C, Naito Y (2012b) In silico polymorphism analysis for the development of simple sequence repeat and transposon markers and construction of linkage map in cultivated peanut. BMC Plant Biol 12(1):80
Shirasawa K, Bhat RS, Khedikar YP, Sujay V, Kolekar RM, Yeri SB, Sukruth M, Cholin S, Asha B, Pandey MK, Fips J (2018) Sequencing analysis of genetic loci for resistance for late leaf spot and rust in peanut (Arachis hypogaea L.). Front Plant Sci 9:1–8
Song GQ, Li MJ, Xiao H, Wang XJ, Tang RH, Xia H, Zhao CZ, Bi YP (2010) EST sequencing and SSR marker development from cultivated peanut (Arachis hypogaea L.). Electron J Biotechnol 13:7–8
Sudhishna E, Srinivas T, Ramesh D, Tushara M (2022) Studies on genetic divergence for yield, yield components and quality traits in peanut (Arachis hypogaea L.). Environ Ecol 40(3):1053–1060
Treutter D (2006) Significance of flavonoids in plant resistance: a review. Environ Chem Lett 4:147–157
Upadhyaya HD (2003) Phenotypic diversity in groundnut (Arachis hypogaea L.) core collection assessed by morphological and agronomical evaluations. Genet Resour Crop Evol 50(5):539–550
Upadhyaya HD, Brame PJ, Ortiz R, Singh S (2002) Developing a mini core of peanut for utilization of genetic resources. Crop Sci 42:2150–2156
Upadhyaya HD, Dwivedi S, Vadez V, Hamidou F, Singh S, Varshney R, Liao B (2014) Multiple resistant and nutritionally dense germplasm identified from mini core collection in peanut. Crop Sci 54:679–693
Waliyar F, Kumar KVK, Diallo M, Traore A, Mangala UN, Upadhyaya HD, Sudini H (2016) Resistance to pre-harvest aflatoxin contamination in ICRISAT’s groundnut mini core collection. Eur J Plant Pathol 145(4):901–913
Wang H, Khera P, Huang B, Yuan M, Katam R, Zhuang W, Harris-Shultz K, Moore KM, Culbreath AK, Zhang X, Varshney RK (2016) Analysis of genetic diversity and population structure of peanut cultivars and breeding lines from China, India and the US using simple sequence repeat markers. Integr Plant Biol 58(5):452–465
Wang H, Zhu S, Dang X, Liu E, Hu X, Eltahawy MS, Zaid IU, Hong D (2019) Favorable alleles mining for gelatinization temperature, gel consistency and amylose content in Oryza sativa by association mapping. BMC Genet 20(1):1–18
Wang CT, Song GS, Wang ZW, Li HJ, Han HW, Chi XY, Wang XZ, Sun XS (2022) Assessment of genetic diversity among Chinese high-oleic peanut genotypes using miniature inverted-repeat transposable element markers. Genet Resour Crop Evol 69(3):949–958
Yang QQ, Cheng L, Long ZY, Li HB, Gunaratne A, Gan RY, Corke H (2019) Comparison of the phenolic profiles of soaked and germinated peanut cultivars via UPLC-QTOF-MS. Antioxidants 8(2):47
Yu B, Jiang H, Pandey MK, Huang L, Huai D, Zhou X, Kang Y, Varshney RK, Sudini HK, Ren X, Luo H (2020) Identification of two novel peanut genotypes resistant to aflatoxin production and their SNP markers associated with resistance. Toxins 12(3):156
Zhang X, Zhang J, He X, Wang Y, Ma X, Yin D (2017) Genome-wide association study of major agronomic traits related to domestication in peanut. Front Plant Sci 8:1611
Zhang L, Qu H, Xie M, Shi T, Shi P, Yu M (2023) Effects of different cooking methods on phenol content and antioxidant activity in sprouted peanut. Molecules 28(12):4684
Zheng Z, Sun Z, Fang Y, Qi F, Liu H, Miao L, Du P, Shi L, Gao W, Han S, Dong W (2018) Genetic diversity, population structure, and botanical variety of 320 global peanut accessions revealed through tunable genotyping-by-sequencing. Sci Rep 8(1):1–10
Zhou Z, Jiang Y, Wang Z, Gou Z, Lyu J, Li W, Yu Y, Shu L, Zhao Y, Ma Y, Fang C (2015) Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat Biotechnol 33(4):408–414
Acknowledgements
The authors acknowledge the funding from the Department of Biotechnology, Government of India to carry out the part of this research and fellowship for SSG.
Funding
The funding was provided by Department of Biotechnology, Government of India (Grant no PR18251, JRF).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gandhadmath, S.S., Vidyashree, S., Choudhary, R. et al. Genetic diversity assessment of groundnut (Arachis hypogaea L.) for polyphenol content and antioxidant activity: unlocking the nutritional potential. J. Plant Biochem. Biotechnol. (2024). https://doi.org/10.1007/s13562-024-00882-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13562-024-00882-4