Abstract
DNA data storage has emerged as a solution for storing massive volumes of data by utilizing nucleic acids as a digital information medium. DNA offers exceptionally high storage density, long durability, and low maintenance costs compared to conventional storage media such as flash memory and hard disk drives. DNA data storage consists of the following steps: encoding, DNA synthesis (i.e., writing), preservation, retrieval, DNA sequencing (i.e., reading), and decoding. Out of these steps, DNA synthesis presents a bottleneck due to imperfect coupling efficiency, low throughput, and excessive use of organic solvents. Overcoming these challenges is essential to establish DNA as a viable data storage medium. In this review, we provide the overall process of DNA data storage, presenting the recent progress of each step. Next, we examine a detailed overview of DNA synthesis methods with an emphasis on their limitations. Lastly, we discuss the efforts to overcome the constraints of each method and their prospects.





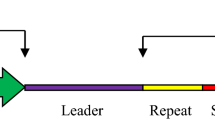
Reprinted with permission from [70]






Similar content being viewed by others
References
Hilbert M, López P. The World’s Technological Capacity to Store, Communicate, and compute information. Science. 2011;332(6025):60–5. https://doi.org/10.1126/science.1200970.
Kim SJ, Jung W-B, Jung HS, Lee M-H, Heo J, Horgan A, Godron X, Ham D. The bottom of the memory hierarchy: Semiconductor and DNA data storage. MRS Bull. 2023;48(5):547–59. https://doi.org/10.1557/s43577-023-00510-x.
Bryan Bishop NM, Victor Zhirnov. Technology Working Group Meeting on future DNA synthesis technologies. Arlington, VA; 2017.
Lunt BM. How Long Is Long-Term Data Storage? Final Program and Proceedings; Conference Location.; 2011:29–33.
Al Kez D, Foley AM, Laverty D, Del Rio DF, Sovacool B. Exploring the sustainability challenges facing digitalization and internet data centers. J Clean Prod. 2022;371; https://doi.org/10.1016/j.jclepro.2022.133633.
Adamcik J, Witz G, Sekatskii SK, Dietler G, Klinov DV. Observation of single-stranded DNA on mica and highly oriented pyrolytic graphite by atomic force microscopy. FEBS Lett. 2006;580(24):5671–5. https://doi.org/10.1016/j.febslet.2006.09.017.
Zhirnov V, Zadegan RM, Sandhu GS, Church GM, Hughes WL. Nucleic acid memory. Nat Mater. 2016;15(4):366–70. https://doi.org/10.1038/nmat4594.
Zhao M, Wen J, Hu Q, Wei X, Zhong Y-W, Ruan H, Gu M. A 3D nanoscale optical disk memory with petabit capacity. Nature. 2024;626(8000):772–8. https://doi.org/10.1038/s41586-023-06980-y.
van der Valk T, Pečnerová P, Díez-del-Molino D, Bergström A, Oppenheimer J, Hartmann S, Xenikoudakis G, Thomas JA, Dehasque M, Sağlıcan E, et al. Million-year-old DNA sheds light on the genomic history of mammoths. Nature. 2021;591(7849):265–9. https://doi.org/10.1038/s41586-021-03224-9.
Baum EB. Building an associative memory vastly larger than the brain. Science. 1995;268(5210):583–5. https://doi.org/10.1126/science.7725109.
Clelland CT, Risca V, Bancroft C. Hiding messages in DNA microdots. Nature. 1999;399(6736):533–4. https://doi.org/10.1038/21092.
Feynman RP. There’s plenty of room at the bottom [data storage]. Journal of Microelectromechanical systems, Microelectromechanical Systems, Journal of. J Microelectromech Syst. 1992;1(1):60–6. https://doi.org/10.1109/84.128057.
Davis J, Microvenus. Art J. 1996;55(1):70. https://doi.org/10.2307/777811.
Church GM, Gao Y, Kosuri S. Next-Generation Digital Information Storage in DNA. Science. 2012;337(6102):1628–1628. https://doi.org/10.1126/science.1226355.
Goldman N, Bertone P, Chen S, Dessimoz C, LeProust EM, Sipos B, Birney E. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature. 2013;494(7435):77–80. https://doi.org/10.1038/nature11875.
Takahashi CN, Nguyen BH, Strauss K, Ceze L. Demonstration of end-to-end automation of DNA Data Storage. Sci Rep. 2019;9:4998. https://doi.org/10.1038/s41598-019-41228-8.
Liu WT, Guo H, Wu JH. Effects of target length on the hybridization efficiency and specificity of rRNA-based oligonucleotide microarrays. Appl Environ Microbiol. 2007;73(1):73. https://doi.org/10.1128/AEM.01468-06. 82-82.
Fan H, Wang J, Komiyama M, Liang X. Effects of secondary structures of DNA templates on the quantification of qPCR. J Biomol Struct Dyn. 2019;37(11):2867–74. https://doi.org/10.1080/07391102.2018.1498804.
Van der Verren SE, Van Gerven N, Jonckheere W, Hambley R, Singh P, Kilgour J, Jordan M, Wallace EJ, Jayasinghe L, Remaut H. A dual-constriction biological nanopore resolves homonucleotide sequences with high fidelity. Nat Biotechnol. 2020. https://doi.org/10.1038/s41587-020-0570-8.
Ross MG, Russ C, Costello M, Hollinger A, Lennon NJ, Hegarty R, Nusbaum C, Jaffe DB. Characterizing and measuring bias in sequence data. Genome Biol. 2013;14(5):1–20. https://doi.org/10.1186/gb-2013-14-5-r51.
Blawat M, Gaedke K, Hütter I, Chen X-M, Turczyk B, Inverso S, Pruitt BW, Church GM. Procedia Comput Sci. 2016;80:1011–22. https://doi.org/10.1016/j.procs.2016.05.398. Forward Error Correction for DNA Data Storage.
Menachem A, Ori DR. An improved Huffman coding method for archiving text, images, and music characters in DNA. Biotechniques. 2009;47(3):747–54. https://doi.org/10.2144/000113218.
Bornhol J, Lopez R, Carmean DM, Ceze L, Seelig G, Strauss K. A DNA-Based archival Storage System. ACM SIGPLAN NOTICES. 2016;51(4):637–49. https://doi.org/10.1145/2872362.2872397.
Reed IS, Solomon G. Polynomial codes over certain Finite fields. J Soc Ind Appl Math. 1960;8(2):300–4.
Agrell E. Errata list for ‘Error Control Coding’ by Lin and Costello. 2011.
Erlich Y, Zielinski D. DNA fountain enables a robust and efficient storage architecture. Science. 2017;355(6328):950–4. https://doi.org/10.1126/science.aaj2038.
MacKay DJC. Fountain codes. IEE Proceedings -- Communications. 2005;152(6):1062–1068; https://doi.org/10.1049/ip-com:20050237.
Wang Y, Noor-A-Rahim M, Zhang J, Gunawan E, Guan YL, Poh CL. High capacity DNA data storage with variable-length oligonucleotides using repeat accumulate code and hybrid mapping. J Biol Eng. 2019;13(1):89. https://doi.org/10.1186/s13036-019-0211-2.
Lee H, Wiegand DJ, Griswold K, Punthambaker S, Chun H, Kohman RE, Church GM. Photon-directed multiplexed enzymatic DNA synthesis for molecular digital data storage. Nat Commun. 2020;11(1):5246. https://doi.org/10.1038/s41467-020-18681-5.
Henry HL, Reza K, Naveen G, Jean B, George MC. Terminator-free template-independent enzymatic DNA synthesis for digital information storage. Nat Commun. 2019;10(1):1–12. https://doi.org/10.1038/s41467-019-10258-1.
Alexander FS, Thuy JDN, Rikke AH, Martin BJ, Troels S, Kurt VG. On-demand synthesis of phosphoramidites. Nat Commun. 2021;12(1):1–7. https://doi.org/10.1038/s41467-021-22945-z.
Schaller H, Weimann G, Lerch B, Khorana HG. Studies on polynucleotides. XXIV. The stepwise synthesis of specific deoxyribopolynucleotides (4). Protected derivatives of Deoxyribonucleosides and new syntheses of Deoxyribonucleoside-3′ phosphates. J Am Chem Soc. 1963;85(23):3821–7. https://doi.org/10.1021/ja00906a021.
Beaucage SL, Caruthers MH. Deoxynucleoside phosphoramidites-A new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 1981;22(20):1859. https://doi.org/10.1016/S0040-4039(01)90461-7. 1862-1862.
Tener G. 2-Cyanoethyl phosphate and its use in the synthesis of phosphate esters. J Am Chem Soc. 1961;83(1):159. https://doi.org/10.1021/ja01462a032. 168-168.
Pirrung MC, Bradley J-C. Comparison of methods for photochemical phosphoramidite-based DNA synthesis. J Org Chem. 1995;60(20):6270. https://doi.org/10.1021/jo00125a010.
Kretschy N, Holik AK, Somoza V, Stengele KP, Somoza MM. Next-generation o-Nitrobenzyl Photolabile groups for Light-Directed Chemistry and microarray synthesis. Angew Chem Int Ed. 2015;54(29):8555–9. https://doi.org/10.1002/anie.201502125.
Grajkowski A, CieŚLak J, Chmielewski MK, MarchÁN V, Phillips LR, Wilk A, Beaucage SL. Conceptual ‘Heat-Driven’ Approach to the synthesis of DNA oligonucleotides on microarrays. Ann N Y Acad Sci. 2003;1002(1):1–11. https://doi.org/10.1196/annals.1281.003.
Septak M. Kinetic studies on depurination and detritylation of CPG-bound intermediates during oligonucleotide synthesis. Nucleic Acids Res. 1996;24(15):3053. https://doi.org/10.1093/nar/24.15.3053. -3058-3058.
Lietard J, Somoza MM, Leger A, Erlich Y, Sadowski N, Timp W. Chemical and photochemical error rates in light-directed synthesis of complex DNA libraries. Nucleic Acids Res. 2021;49(12):6687–701. https://doi.org/10.1093/nar/gkab505.
Vargeese C, Carter J, Yegge J, Krivjansky S, Settle A, Kropp E, Peterson K, Pieken W. Efficient activation of nucleoside phosphoramidites with 4,5-dicyanoimidazole during oligonucleotide synthesis. Nucleic Acids Res. 1998;26(4):1046–50. https://doi.org/10.1093/nar/26.4.1046.
Caruthers MH. The Chemical synthesis of DNA/RNA: our gift to Science. J Biol Chem. 2013;288(2):1420–7. https://doi.org/10.1074/jbc.X112.442855.
Beaucage SL. Strategies in the preparation of DNA oligonucleotide arrays for diagnostic applications. Curr Med Chem. 2001;8(10):1213–44. https://doi.org/10.2174/0929867013372463.
LeProust E. Characterization of oligodeoxyribonucleotide synthesis on glass plates. Nucleic Acids Res. 2001;29(10):2171. https://doi.org/10.1093/nar/29.10.2171.
David MJ, Lilley MGFE. Who will fill the gap by making nucleic synthesizers now? Nature. 2001;411(6833):15–15. https://doi.org/10.1038/35075244.
Jobs M, Fredrikkson S, Brookes AJ, Landegren U. Effect of Oligonucleotide truncation on single-nucleotide distinction by solid-phase hybridization. Anal Chem. 2002;74(1):199. https://doi.org/10.1021/ac010555s.
Blanchard AP, Hood L. Sequence to array: probing the genome’s secrets. Nat Biotechnol. 1996;14(12):1649–1649. https://doi.org/10.1038/nbt1296-1649.
LeProust EM, Peck BJ, Spirin K, McCuen HB, Moore B, Namsaraev E, Caruthers MH. Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process. Nucleic Acids Res. 2010;38(8):2522–40. https://doi.org/10.1093/nar/gkq163.
Tjong V, Yu H, Hucknall A, Rangarajan S, Chilkoti A. Amplified on-chip fluorescence detection of DNA hybridization by surface-initiated enzymatic polymerization. Anal Chem. 2011;83(13):5153–9. https://doi.org/10.1021/ac200946t.
Tang L, Zauscher S, Tjong V, Chilkoti A, Li N, Yingling YG. Enzymatic polymerization of high molecular weight DNA amphiphiles that self-assemble into star-like micelles. Adv Mater. 2014;26(19):3050–4. https://doi.org/10.1002/adma.201306049.
Tang L, Navarro LA, Zauscher S, Chilkoti A. High-molecular-weight polynucleotides by transferase-catalyzed living chain-growth polycondensation. Angewandte Chemie - Int Ed. 2017;56(24):6778–82. https://doi.org/10.1002/anie.201700991.
Mackey JK, Gilham PT. New Approach to the synthesis of polyribonucleotides of defined sequence. Nature. 1971;233(5321):551–3. https://doi.org/10.1038/233551a0.
Hinton DM, Gumport RI. The synthesis of oligodeoxyribonucleotides using RNA ligase. Nucleic Acids Res. 1979;7(2):453–64. https://doi.org/10.1093/nar/7.2.453.
Hoff K, Halpain M, Garbagnati G, Edwards JS, Zhou W. Enzymatic synthesis of designer DNA using cyclic reversible termination and a Universal Template. ACS Synth Biol. 2020;9(2):283. https://doi.org/10.1021/acssynbio.9b00315. 293-293.
Gouge J, Rosario S, Romain F, Poitevin F, Béguin P, Delarue M. Structural basis for a novel mechanism of DNA bridging and alignment in eukaryotic DSB DNA repair. EMBO J. 2015;34(8):1126–42. https://doi.org/10.15252/embj.201489643.
Loc’h J, Delarue M. Terminal deoxynucleotidyltransferase: the story of an untemplated DNA polymerase capable of DNA bridging and templated synthesis across strands. Curr Opin Struct Biol. 2018;53:22–31. https://doi.org/10.1016/j.sbi.2018.03.019.
Verardo D, Adelizzi B, Rodriguez-Pinzon DA, Moghaddam N, Thomée E, Loman T, Godron X, Horgan A. Multiplex enzymatic synthesis of DNA with single-base resolution. Sci Adv. 2023;9(27). https://doi.org/10.1126/sciadv.adi0263.
Palluk S, Arlow DH, de Rond T, Barthel S, Kang JS, Bector R, Baghdassarian HM, Truong AN, Kim PW, Singh AK et al. De novo DNA synthesis using polymerase-nucleotide conjugates. Nat Biotechnol. 2018(7);.
Ju J, Kim DH, Bi L, Meng Q, Bai X, Li Z, Li X, Marma MS, Shi S, Wu J, et al. Proc Natl Acad Sci USA. 2006;103(52):19635–40. https://doi.org/10.1073/pnas.0609513103. Four-Color DNA Sequencing by Synthesis Using Cleavable Fluorescent Nucleotide Reversible Terminators.
Mayer C, McInroy GR, Murat P, Van Delft P, Balasubramanian S. An epigenetics-inspired DNA-Based data Storage System. Angew Chem Int Ed. 2016;55(37):11144–8. https://doi.org/10.1002/anie.201605531.
Ren Y, Zhang Y, Liu Y, Wu Q, Su J, Wang F, Chen D, Fan C, Liu K, Zhang H. Small Methods. 2022;6(4):1–9. https://doi.org/10.1002/smtd.202101335. DNA-Based Concatenated Encoding System for High‐Reliability and High‐Density Data Storage.
Tabatabaei SK, Pham B, Pan C, Liu J, Chandak S, Shorkey SA, Hernandez AG, Aksimentiev A, Chen M, Schroeder CM, Milenkovic O. Nano Lett. 2022;22(5):1905–14. https://doi.org/10.1021/acs.nanolett.1c04203. Expanding the Molecular Alphabet of DNA-Based Data Storage Systems with Neural Network Nanopore Readout.
Yeongjae C, Taehoon R, Amos CL, Hansol C, Hansaem L, Jaejun P, Suk-Heung S, Seojoo K, Hyeli K, Wook P, Sunghoon K. High information capacity DNA-based data storage with augmented encoding characters using degenerate bases. Sci Rep. 2019;9(1):1–7. https://doi.org/10.1038/s41598-019-43105-w.
Nathaniel R, Swapnil PB, Sarah AF, Sean M, Michael WN, Devin L, Hyunjun P. DNA-based data storage via combinatorial assembly. bioRxiv. 2021:2021.2004.2020.440194; https://doi.org/10.1101/2021.04.20.440194.
Randolph L, Yuan-Jyue C, Siena Dumas A, Sergey Y, Konstantin M, Miklos ZR, Georg S, Karin S, Luis C. DNA assembly for nanopore data storage readout. Nat Commun. 2019;10(1):1–9. https://doi.org/10.1038/s41467-019-10978-4.
Li S-Y, Liu J-K, Zhao G-P, Wang J. CADS: CRISPR/Cas12a-Assisted DNA steganography for securing the storage and transfer of DNA-Encoded information. ACS Synth Biol. 2018;7(4):1174–8. https://doi.org/10.1021/acssynbio.8b00074.
Shipman SL, Nivala J, Macklis JD, Church GM. CRISPR-Cas encoding of a digital movie into the genomes of a population of living bacteria. Nature. 2017;547(7663):345–9. https://doi.org/10.1038/nature23017.
Chen W, Han M, Zhou J, Ge Q, Wang P, Zhang X, Zhu S, Song L, Yuan Y. An artificial chromosome for data storage. Natl Sci Rev. 2021;8(5). https://doi.org/10.1093/nsr/nwab028.
McNally B, Singer A, Yu Z, Sun Y, Weng Z, Meller A. Optical recognition of converted DNA nucleotides for single-molecule DNA sequencing using nanopore arrays. Nano Lett. 2010;10(6):2237–44. https://doi.org/10.1021/nl1012147.
Baek D, Joe S-Y, Shin H, Park C, Jo S, Chun H. Recent progress in high-throughput enzymatic DNA synthesis for Data Storage. Biochip J. 2024. https://doi.org/10.1007/s13206-024-00146-2.
Choi Y, Bae HJ, Lee AC, Choi H, Lee D, Ryu T, Hyun J, Kim S, Kim H, Song SH, et al. DNA micro-disks for the management of DNA‐Based Data Storage with Index and write‐once–read‐many (WORM) memory features. Adv Mater. 2020;32(37):1–8. https://doi.org/10.1002/adma.202001249.
Thompson B, Bundell S. A mammoth discovery: oldest DNA on record from million-year-old teeth. Nature. 2021. https://doi.org/10.1038/d41586-021-00442-z.
Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362(6422):709. https://doi.org/10.1038/362709a0.
Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11(19):3610–8. https://doi.org/10.1021/bi00769a018.
Tubbs A, Nussenzweig A. Endogenous DNA damage as a source of genomic instability in Cancer. Cell. 2017;168(4):644–56. https://doi.org/10.1016/j.cell.2017.01.002.
Kaplan M. DNA has a 521-year half-life. Nature. 2023;Preprints. https://doi.org/10.1038/nature.2012.11555.
DNA legacy. Accessed 03 Jan 2023.
Paunescu D, Puddu M, Soellner JOB, Stoessel PR, Grass RN. Reversible DNA encapsulation in silica to produce ROS-resistant and heat-resistant synthetic DNA ‘fossils’. Nat Protoc. 2013;8(12):2440–8. https://doi.org/10.1038/nprot.2013.154.
Bonnet J, Colotte M, Coudy D, Couallier V, Portier J, Morin B, Tuffet S. Chain and conformation stability of solid-state DNA: implications for room temperature storage. Nucleic Acids Res. 2010;38(5):1531–46. https://doi.org/10.1093/nar/gkp1060.
Matthew D, James L, C L, M AJ, Eske, Guojie PR. Michael. The half-life of DNA in bone: measuring decay kinetics in 158 dated fossils. Proceedings of the Royal Society B: Biological Sciences. 2012;279(1748):4724–4733; https://doi.org/10.1098/rspb.2012.1745.
Koch J, Gantenbein S, Masania K, Stark WJ, Erlich Y, Grass RN. A DNA-of-things storage architecture to create materials with embedded memory. Nat Biotechnol. 2020;38(1):39–. https://doi.org/10.1038/s41587-019-0356-z.
Liu Q, Wei Y, Wang Z, Song DP, Cui J, Qi H, Sustainable. Small Methods. 2023;7(9):1–9. https://doi.org/10.1002/smtd.202201610. DNA Data Storage on Cellulose Paper.
Liu Y, Zheng Z, Gong H, Liu M, Guo S, Li G, Wang X, Kaplan DL. DNA preservation in silk. Biomaterials Science (2047–4830). 2017;5(7):1279–1292; https://doi.org/10.1039/c6bm00741d.
Kohll AX, Antkowiak PL, Chen WD, Nguyen BH, Stark WJ, Ceze L, Strauss K, Grass RN. Stabilizing synthetic DNA for long-term data storage with earth alkaline salts. Chem Commun. 2020;56(25):3613–6. https://doi.org/10.1039/d0cc00222d.
Antkowiak PL, Koch J, Rzepka P, Nguyen BH, Strauss K, Stark WJ, Grass RN. Anhydrous calcium phosphate crystals stabilize DNA for dry storage. Chem Commun. 2022;58(19):3174–7. https://doi.org/10.1039/d2cc00414c.
Paulsen KS, Di Carlo D, Chung AJ. Optofluidic fabrication for 3D-shaped particles. Nat Commun. 2015;6:6976. https://doi.org/10.1038/ncomms7976.
Kozarewa I, Armisen J, Gardner AF, Slatko BE, Hendrickson CL. Overview of Target Enrichment Strategies. Current protocols in molecular biology. 2015;112:7.21.21–27.21.23; https://doi.org/10.1002/0471142727.mb0721s112.
Tomek KJ, Volkel K, Simpson A, Hass AG, Indermaur EW, Tuck JM, Keung AJ. Driving the scalability of DNA-Based information Storage systems. ACS Synth Biol. 2019;8(6):1241–8. https://doi.org/10.1021/acssynbio.9b00100.
Grass RN, Heckel R, Puddu M, Paunescu D, Stark WJ. Robust Chemical Preservation of Digital Information on DNA in silica with error-correcting codes. Angew Chem Int Ed. 2015;54(8):2552–5. https://doi.org/10.1002/anie.201411378.
Paunescu D, Fuhrer R, Grass RN. Protection and Deprotection of DNA-High-Temperature Stability of Nucleic Acid barcodes for Polymer labeling. Angew Chem Int Ed. 2013;52(15):4269–72. https://doi.org/10.1002/anie.201208135.
Puddu M, Stark WJ, Grass RN. Silica microcapsules for Long-Term, Robust, and Reliable Room temperature RNA preservation. Adv Healthc Mater. 2015;4(9):1332–8. https://doi.org/10.1002/adhm.201500132.
Chen WD, Kohll AX, Nguyen BH, Koch J, Heckel R, Stark WJ, Ceze L, Strauss K, Grass RN. Combining data longevity with high Storage Capacity-Layer-by-layer DNA encapsulated in magnetic nanoparticles. Adv Funct Mater. 2019;29(28):1901672. https://doi.org/10.1002/adfm.201901672.
Banal JL, Shepherd TR, Berleant J, Huang H, Reyes M, Ackerman CM, Blainey PC, Bathe M. Random access DNA memory using boolean search in an archival file storage system. Nat Mater. 2021;20(9):1272–. https://doi.org/10.1038/s41563-021-01021-3.
Antkowiak PL, Koch J, Nguyen BH, Stark WJ, Strauss K, Ceze L, Grass RN. Integrating DNA encapsulates and Digital Microfluidics for Automated Data Storage in DNA. Small. 2022;18:1–9. https://doi.org/10.1002/smll.202107381.
Stephenson A, Willsey M, McBride J, Newman S, Nguyen B, Takahashi C, Strauss K, Ceze L. PurpleDrop: A Digital Microfluidics-Based Platform for Hybrid Molecular-Electronics Applications. IEEE Micro, Micro, IEEE. 2020;40(5):76–86; https://doi.org/10.1109/MM.2020.3005615.
Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475(7356):348–52. https://doi.org/10.1038/nature10242.
Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74(12):5463–7. https://doi.org/10.1073/pnas.74.12.5463.
Check Hayden E, Technology. The $1,000 genome. Nature. 2014;507(7492):294–5. https://doi.org/10.1038/507294a.
Slatko BE, Gardner AF, Ausubel FM. Overview of next-generation sequencing technologies. Curr Protoc Mol Biol. 2018;122(1):e59. https://doi.org/10.1002/cpmb.59.
Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol. 2000;18(6):630. https://doi.org/10.1038/76469.
Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11(1):31–46. https://doi.org/10.1038/nrg2626.
Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, Vollger MR, Altemose N, Uralsky L, Gershman A, et al. The complete sequence of a human genome. Science. 2022;376(6588):44–53. https://doi.org/10.1126/science.abj6987.
Katoh K, Standley DM. MAFFT multiple sequence alignment Software Version 7: improvements in performance and usability. Mol Biology Evol. 2013;30(4):772–80. https://doi.org/10.1093/molbev/mst010.
Xie R, Zan X, Chu L, Su Y, Xu P, Liu W. Study of the error correction capability of multiple sequence alignment algorithm (MAFFT) in DNA storage. BMC Bioinformatics. 2023;24(1):1–11. https://doi.org/10.1186/s12859-023-05237-9.
Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26(10):1146–53. https://doi.org/10.1038/nbt.1495.
Dohm JC, Peters P, Stralis-Pavese N, Himmelbauer H. Benchmarking of long-read correction methods. NAR Genomics Bioinf. 2020;2(2):lqaa037. https://doi.org/10.1093/nargab/lqaa037.
Metzker ML. Sequencing in real time. Nat Biotechnol. 2009;27(2):150–1. https://doi.org/10.1038/nbt0209-150.
Levene MJ, Korlach J, Turner SW, Foquet M, Craighead HG, Webb WW. Zero-Mode Waveguides for single-molecule analysis at high concentrations. Science. 2003;299(5607):682–6. https://doi.org/10.1126/science.1079700.
Illumina novaseq 6000. Accessed 03 Jan 2023.
Oxford Nanopore MinIon. Accessed 03 Jan 2024.
Jain M, Fiddes IT, Miga KH, Olsen HE, Paten B, Akeson M. Improved data analysis for the MinION nanopore sequencer. Nat Methods. 2015;12(4):351–6. https://doi.org/10.1038/nmeth.3290.
Hon T, Mars K, Young G, Tsai Y-C, Karalius JW, Landolin JM, Maurer N, Kudrna D, Hardigan MA, Steiner CC, et al. Highly accurate long-read HiFi sequencing data for five complex genomes. Sci Data. 2020;7(1):399. https://doi.org/10.1038/s41597-020-00743-4.
Jiawen C, Nan s, Zhaogeng L, Guolu X, Yuyao W, Biao J. Analysis and comprehensive comparison of PacBio and nanopore-based RNA sequencing of the Arabidopsis transcriptome. Plant Methods. 2020;16(1):1–13. https://doi.org/10.1186/s13007-020-00629-x.
Pac Bio Sequel. Accessed 03 Jan 2023.
Thermo Fisher Ion Torrent S5. Accessed 03 Jan 2024.
Carlson R. The changing economics of DNA synthesis. Nat Biotechnol. 2009;27(12):1091–4. https://doi.org/10.1038/nbt1209-1091.
Hoose A, Vellacott R, Storch M, Freemont PS, Ryadnov MG. DNA synthesis technologies to close the gene writing gap. Nat Reviews Chem. 2023;7(3):144–61. https://doi.org/10.1038/s41570-022-00456-9.
Dalma-Weiszhausz DD, Warrington J, Tanimoto EY, Miyada CG. [1] the Affymetrix GeneChip® platform: an overview. Methods Enzymol. 2006;410:3–28. https://doi.org/10.1016/S0076-6879(06)10001-4.
Jingdong T, Hui G, Nijing S, Xiaochuan Z, Erdogan G, Xiaolian G, Church G. Accurate multiplex gene synthesis from programmable DNA microchips. Nature. 2004;432(7020):1050–4. https://doi.org/10.1038/nature03151.
Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, Lefkowitz SM, Ziman M, Schelter JM, Meyer MR, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol. 2001;19(4):342. https://doi.org/10.1038/86730.
Blanchard AP, Kaiser RJ, Hood LE. High-density oligonucleotide arrays. Biosens Bioelectron. 1996;11(6):687–90. https://doi.org/10.1016/0956-5663(96)83302-1.
Pease AC, Solas D, Sullivan EJ, Cronin MT, Holmes CP, Fodor SP. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc Natl Acad Sci USA. 1994;91(11):5022–6. https://doi.org/10.1073/pnas.91.11.5022.
Barone A, Beecher J, Bury P, Chen C, Doede T, Fidanza J, McGall G. Photolithographic synthesis of high-desity oligonucleotide probe arrays. Nucleosides Nucleotides Nucleic Acids. 2001;20(4–7):525–31. https://doi.org/10.1081/NCN-100002328.
Beier M, Hoheisel JD. Production by quantitative photolithographic synthesis of individually quality checked DNA microarrays. Nucleic Acids Res. 2000;28(4):E11. https://doi.org/10.1093/nar/28.4.e11.
McGall GH, Barone AD, Diggelmann M, Fodor SPA, Gentalen E, Ngo N. The efficiency of light-directed synthesis of DNA arrays on glass substrates. J Am Chem Soc. 1997;119(22):5081. https://doi.org/10.1021/ja964427a.
Pirrung MC, Fallon L, McGall G. Proofing of photolithographic DNA synthesis with 3’,5’-dimethoxybenzoinyloxycarbonyl-protected. J Org Chem. 1998;63(2):241. https://doi.org/10.1021/jo970872s.
McGall G, Labadie J. Light-directed synthesis of high-density oligonucleotide arrays using semiconductor photoresists. Proc Natl Acad Sci USA. 1996;93(24):13555. https://doi.org/10.1073/pnas.93.24.13555.
Hornbeck LJ. Digital Light Processing and MEMS: reflecting the digital display needs of the networked society. Proceedings of SPIE. 1996(1):2–13; https://doi.org/10.1117/12.248477.
Philipp LA, Jory L, Mohammad Zalbagi D, Mark MS, Wendelin JS, Reinhard H, Robert NG. Low cost DNA data storage using photolithographic synthesis and advanced information reconstruction and error correction. Nat Commun. 2020;11(1):1–10. https://doi.org/10.1038/s41467-020-19148-3.
Agbavwe C, Kim C, Hong D, Heinrich K, Wang T, Somoza MM, Efficiency. Error and yield in Light-Directed Maskless synthesis of DNA microarrays. J Nanobiotechnol. 2011;9(1):57–73. https://doi.org/10.1186/1477-3155-9-57.
LeProust E, Pellois JP, Yu P, Zhang H, Gao X, Srivannavit O, Gulari E, Zhou X. Digital light-directed synthesis. A microarray platform that permits rapid reaction optimization on a combinatorial basis. J Comb Chem. 2000;2(4):349–54. https://doi.org/10.1021/cc000009x.
Gao X, LeProust E, Zhang H, Srivannavit O, Gulari E, Yu P, Nishiguchi C, Xiang Q, Zhou X. A flexible light-directed DNA chip synthesis gated by deprotection using solution photogenerated acids. Nucleic Acids Res. 2001;29(22):4744–50. https://doi.org/10.1093/nar/29.22.4744.
Singh-Gasson S, Green RD, Yue Y, Nelson C, Blattner F, Sussman MR, Cerrina F. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat Biotechnol. 1999;17(10):974. https://doi.org/10.1038/13664.
Egeland RD, Marken F, Southern EM. An Electrochemical Redox couple activated by microelectrodes for Confined Chemical patterning of surfaces. Anal Chem. 2002;74(7):1590. https://doi.org/10.1021/ac010953v.
Egeland RD, Southern EM. Electrochemically directed synthesis of oligonucleotides for DNA microarray fabrication. Nucleic Acids Res. 2005;33(14):e125. https://doi.org/10.1093/nar/gni117.
Ghindilis AL, Smith MW, Schwarzkopf KR, Roth KM, Peyvan K, Munro SB, Lodes MJ, Stöver AG, Bernards K, Dill K, McShea A. CombiMatrix oligonucleotide arrays: genotyping and gene expression assays employing electrochemical detection. Biosens Bioelectron. 2007;22(9):1853–60. https://doi.org/10.1016/j.bios.2006.06.024.
Nguyen BH, Takahashi CN, Gupta G, Smith JA, Rouse R, Berndt P, Yekhanin S, Ward DP, Ang SD, Garvan P, et al. Scaling DNA data storage with nanoscale electrode wells. Sci Adv. 2021;7(48):1–6. https://doi.org/10.1126/sciadv.abi6714.
Lausted C, Dahl T, Warren C, King K, Smith K, Johnson M, Saleem R, Aitchison J, Hood L, Lasky SR. POSaM: a fast, flexible, open-source, inkjet oligonucleotide synthesizer and microarrayer. Genome Biol. 2004;5(8):R58. https://doi.org/10.1186/gb-2004-5-8-r58.
Chow BY, Emig CJ, Jacobson JM. Photoelectrochemical synthesis of DNA microarrays. Proc Natl Acad Sci USA. 2009;106(36):15219–24. https://doi.org/10.1073/pnas.0813011106.
Andrew JF, Matthew JH, Blair CK, Yen-chun L, Vijay N, Albert P. Thermofluidic chip containing virtual thermal wells. Eng Biology. 2019. https://doi.org/10.1049/enb.2018.5010.
Gao X, Yu P, LeProust E, Sonigo L, Pellois JP, Zhang H. Oligonucleotide synthesis using solution photogenerated acids [17]. J Am Chem Soc. 1998;120(48):12698–9. https://doi.org/10.1021/ja9827191.
Chun H. Highly precision DNA photosynthesis based on self-aligned micropattern by image reversal. PCT/KR2023/004979; 2023.
Karl Maurer JJC Jr., Fuji HS. Joseph Leonetti. Neutralization and containment of redox species produced by circumferential electrodes. US. 2021;11(838):B2.
Srivannavit O, Gulari M, Gulari E, LeProust E, Pellois JP, Gao X, Zhou X. Design and fabrication of microwell array chips for a solution-based, photogenerated acid-catalyzed parallel oligonuclotide DNA synthesis. Sens Actuators: Phys. 2004;116(1):150–60. https://doi.org/10.1016/j.sna.2004.04.025.
Karl Maurer JC, Marcelo Caraballo,James Crye,Dominic Suciu,Andrey Ghindilis,Joseph, Leonetti,Wei A, Wang FM, Rossi,Axel G. Stöver,Christopher Larson,Hetian Gao,Kilian Dill,Andy McShea. Electrochemically Generated Acid and Its Containment to 100 Micron Reaction Areas for the Production of DNA Microarrays. PLoS ONE. 2006:1–7; https://doi.org/10.1371/journal.pone.0000034.
William Banyai BJP, Fernandez A, Chen S. Pierre Indermuhle. DE NOVO SYNTHESIZED GENE LIBRARIES. US 9,555,388 B2; 2017.
Carr PA, Church GM. Genome engineering. Nat Biotechnol. 2009;27(12):1151–62. https://doi.org/10.1038/nbt.1590.
Agarwal KL, BÜChi H, Caruthers MH, Gupta N, Khorana HG, Kleppe K, Kumar A, Ohtsuka E, Rajbhandary UL, Van De Sande JH, et al. Total synthesis of the gene for an alanine transfer ribonucleic acid from yeast. Nature. 1970;227(5253):27–34. https://doi.org/10.1038/227027a0.
Stemmer WPC, Crameri A, Ha KD, Brennan TM, Heyneker HL. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164(1):49. https://doi.org/10.1016/0378-1119(95)00511-4. 53-53.
Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–5. https://doi.org/10.1038/nmeth.1318.
SurePrint G. CGH & CGH + SNP Microarrays. Accessed 8 Nov 2023.
Twist Bioscience. Accessed 07 Jan 2024.
Nimblegen. Accessed 07 Jan 2024.
Miniature Semiconductor Technology for Nucleic Acid Synthesis. Accessed 8 Nov 2023.
Smith JA, Nguyen BH, Carlson R, Bertram JG, Palluk S, Arlow DH, Strauss K. Spatially selective Electrochemical cleavage of a polymerase-nucleotide conjugate. ACS Synth Biol. 2023;12(6):1716–26. https://doi.org/10.1021/acssynbio.3c00044.
Jung HS, Jung W-B, Wang J, Abbott J, Horgan A, Fournier M, Hinton H, Hwang Y-H, Godron X, Nicol R, et al. CMOS electrochemical pH localizer-imager. Sci Adv. 2022;8(30):eabm6815. https://doi.org/10.1126/sciadv.abm6815.
Verardo D, Adelizzi B, Rodriguez-Pinzon DA, Moghaddam N, Thomée E, Loman T, Godron X, Horgan A. Multiplex enzymatic synthesis of DNA with single-base resolution. Sci Adv. 2023;9(27):eadi0263. https://doi.org/10.1126/sciadv.adi0263.
Pham TM, Tan KW, Sakumura Y, Okumura K, Maki H, Akiyama MT. A single-molecule approach to DNA replication in Escherichia coli cells demonstrated that DNA polymerase III is a major determinant of fork speed. Mol Microbiol. 2013;90(3):584–96. https://doi.org/10.1111/mmi.12386.
Boeke JD, Church G, Hessel A, Kelley NJ, Arkin A, Yizhi C, Carlson R, Chakravarti A, Cornish VW, Holt L, et al. The Genome Project–write. Science. 2016;353(6295):126–7. https://doi.org/10.1126/science.aaf6850.
Vitak S. Technology alliance boosts efforts to store data in DNA. Nature. 2021. https://doi.org/10.1038/d41586-021-00534-w.
Funding
This work was supported by the National Research Foundation of Korea (NRF) (NRF-2020R1A2C3010322).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design as well as manuscript preparation. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jo, S., Shin, H., Joe, Sy. et al. Recent progress in DNA data storage based on high-throughput DNA synthesis. Biomed. Eng. Lett. (2024). https://doi.org/10.1007/s13534-024-00386-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13534-024-00386-z