Abstract
Purpose
Urothelial carcinoma (UC) is a common disease in developed counties. This study aimed to identify autocrine roles and signaling pathways of gremlin 1, DAN family BMP antagonist (GREM1), which inhibits tumor growth and epithelial-mesenchymal transition (EMT) in UC.
Methods
Systematic in vitro and in vivo studies using genetic engineering, different urinary bladder urothelial carcinoma (UBUC)-derived cell lines, and mouse models were performed, respectively. Further, primary upper tract urothelial carcinoma (UTUC) and UBUC specimens were evaluated by immunohistochemistry.
Results
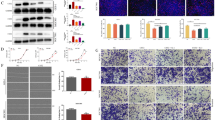
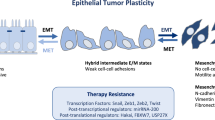
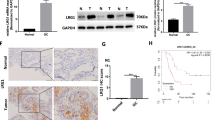
GREM1 protein levels conferred better disease-specific and metastasis-free survival rates and played an independent prognostic factor in UTUC and UBUC. Hypermethylation is the primary cause of low GREM1 levels. In different UBUC-derived cell lines, the autocrine/secreted and glycosylated GREM1 interacted with transforming growth factor beta 1 (TGFB1) and inhibited TGFβ/BMP/SMAD signaling and myosin light chain 9 (MYL9) transactivation, subsequently cell proliferation and epithelial-mesenchymal transition (EMT). Secreted and glycosylated GREM1 also suppressed tumor growth, metastasis, and MYL9 levels in the mouse model. Instead, cytosolic GREM1 promoted cell proliferation and EMT by activating the tumor necrosis factor (TNF)/AKT/nuclear factor kappa B (NFκB) axis.
Conclusions
Clinical associations, animal models, and in vitro indications provided solid evidence to show that the epithelial autocrine GREM1 is a novel tumor suppressor in UCs. The glycosylated-GREM1 hampered cell proliferation, migration, invasion, and in vitro angiogenesis through interaction with TGFB1 to inactivate TGFβ/BMP/SMAD-mediated EMT in an autocrine manner.









Similar content being viewed by others
Data availability
The raw data used to support the conclusions of this article will be made available by the corresponding author, without undue reservation to any qualified researcher.
Abbreviations
- 5-aza :
-
5-Aza-2’-deoxycytidine
- AKT1 :
-
AKT serine/threonine kinase 1
- AMH :
-
Anti-Mullerian hormone
- bHLHs :
-
Basic HLHs
- BLCA :
-
Bladder urothelial carcinoma
- BMP :
-
Bone morphogenetic protein
- BMPR2 :
-
Bone morphogenetic protein receptor type 2
- BrdU :
-
5-Bromo-2'-deoxyuridine
- BRE :
-
BMP responsive element
- BTRC :
-
Beta-transducin repeat containing E3 ubiquitin protein
- CDH1 :
-
Cadherin 1
- CDKN1A :
-
Cyclin-dependent kinase inhibitor 1 A
- DAPK1 :
-
Death-associated protein kinase 1
- DSS :
-
Disease-specific survival
- EMT :
-
Epithelial-mesenchymal transition
- FN1 :
-
Fibronectin 1
- GDF :
-
Growth differentiation factors
- GEO :
-
Gene Expression Omnibus
- GREM1 :
-
Gremlin 1, DAN family BMP antagonist
- GSEA :
-
Gene Set Enrichment Analysis
- HUVEC :
-
Human umbilical vein endothelial cells
- ID1 :
-
Inhibitor of DNA binding 1
- IKBKB :
-
Nuclear factor kappa B kinase subunit beta
- MFS :
-
Metastasis-free survival
- MGMT :
-
O-6-methylguanine-DNA methyltransferase
- MLH1 :
-
MutL homolog 1
- MMP :
-
Matrix metalloproteinase
- MTT :
-
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- MYL9 :
-
Myosin light chain 9
- NFκB :
-
Nuclear factor kappa B
- NFKBIA :
-
NFKB inhibitor alpha
- NGF :
-
Nerve growth factor
- NODAL :
-
Nodal growth differentiation factor
- PDAC :
-
Pancreatic ductal adenocarcinoma
- PDGF :
-
Platelet-derived growth factor
- PI3K :
-
Phosphoinositide 3-kinase
- PIK3CA :
-
Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha
- RELA :
-
RELA proto-oncogene
- RPKM :
-
Read per kilobase per million
- shRNA :
-
Small hairpin RNA
- SNAI1 :
-
Snail family transcriptional repressor 1
- SRE :
-
SMAD responsive element
- TCGA :
-
The Cancer Genome Atlas
- TGFβ :
-
Transforming growth factor beta
- TGFBR :
-
Transforming growth factor beta receptor
- TNF :
-
Tumor necrosis factor
- TNFRSF1B :
-
TNF receptor superfamily member 1B
- TP53 :
-
Tumor protein p53
- TRAF1 :
-
TNF receptor-associated factor 1
- TWIST1 :
-
Twist family bHLH transcription factor 1
- UBUC :
-
Urinary bladder urothelial carcinoma
- UC :
-
Urothelial carcinoma
- UTUC :
-
Upper tract urothelial carcinoma
- VIM :
-
Vimentin
- ZEB1 :
-
Zinc finger E-box binding homeobox 1
References
O. Sanli, J. Dobruch, M.A. Knowles, M. Burger, M. Alemozaffar, M.E. Nielsen, Y. Lotan, Bladder cancer. Nat. Rev. Dis. Primers 3, 17022 (2017)
M. Rouprêt, M. Babjuk, M. Burger, O. Capoun, D. Cohen, E.M. Compérat, N.C. Cowan, J.L. Dominguez-Escrig, P. Gontero, A. Hugh Mostafid et al., European association of urology guidelines on upper urinary tract urothelial carcinoma: 2020 update. Eur. Urol. 79(1), 62–79 (2021)
K.K. Aben, J.A. Witjes, M.P. Schoenberg, C. Hulsbergen-van de Kaa, A.L. Verbeek, L.A. Kiemeney, Familial aggregation of urothelial cell carcinoma. Int. J. Cancer 98(2), 274–278 (2002)
P. Lichtenstein, N.V. Holm, P.K. Verkasalo, A. Iliadou, J. Kaprio, M. Koskenvuo, E. Pukkala, A. Skytthe, K. Hemminki, Environmental and heritable factors in the causation of cancer–analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 343(2), 78–85 (2000)
T. Powles, J. Bellmunt, E. Comperat, M. De Santis, R. Huddart, Y. Loriot, A. Necchi, B.P. Valderrama, A. Ravaud, S.F. Shariat et al., Bladder cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 33(3), 244–258 (2022)
A. Richters, K.K.H. Aben, L. Kiemeney, The global burden of urinary bladder cancer: an update. World J. Urol. 38(8), 1895–1904 (2020)
J.J. Meeks, H. Al-Ahmadie, B.M. Faltas, J.A. Taylor 3rd., T.W. Flaig, D.J. DeGraff, E. Christensen, B.L. Woolbright, D.J. McConkey, L. Dyrskjøt, Genomic heterogeneity in bladder cancer: challenges and possible solutions to improve outcomes. Nat. Rev. Urol. 17(5), 259–270 (2020)
L. Fagerberg, B.M. Hallström, P. Oksvold, C. Kampf, D. Djureinovic, J. Odeberg, M. Habuka, S. Tahmasebpoor, A. Danielsson, K. Edlund et al., Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics 13(2), 397–406 (2014)
X. Guo, X.F. Wang, Signaling cross-talk between TGF-beta/BMP and other pathways. Cell. Res. 19(1), 71–88 (2009)
L.Z. Topol, B. Bardot, Q. Zhang, J. Resau, E. Huillard, M. Marx, G. Calothy, D.G. Blair, Biosynthesis, post-translation modification, and functional characterization of Drm/Gremlin. J. Biol. Chem. 275(12), 8785–8793 (2000)
UniProt, UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 49(D1), D480-d489 (2021)
G.M. Todd, Z. Gao, M. Hyvönen, D.P. Brazil, P. Ten Dijke, Secreted BMP antagonists and their role in cancer and bone metastases. Bone 137, 115455 (2020)
D.P. Brazil, R.H. Church, S. Surae, C. Godson, F. Martin, BMP signalling: agony and antagony in the family. Trends Cell Biol. 25(5), 249–264 (2015)
S. O’Reilly, Gremlin: a complex molecule regulating wound healing and fibrosis. Cell. Mol. Life Sci. 78(24), 7917–7923 (2021)
R.H. Church, A. Krishnakumar, A. Urbanek, S. Geschwindner, J. Meneely, A. Bianchi, B. Basta, S. Monaghan, C. Elliot, M. Strömstedt et al., Gremlin1 preferentially binds to bone morphogenetic protein-2 (BMP-2) and BMP-4 over BMP-7. Biochem. J. 466(1), 55–68 (2015)
I.J.H. van Vlodrop, S.C. Joosten, T. De Meyer, K.M. Smits, L. Van Neste, V. Melotte, M. Baldewijns, L.J. Schouten, P.A. van den Brandt, J. Jeschke et al., A four-gene promoter methylation marker panel consisting of GREM1, NEURL, LAD1, and NEFH predicts survival of clear cell renal cell cancer patients. Clin. Cancer Res. 23(8), 2006–2018 (2017)
M.R. Morris, C. Ricketts, D. Gentle, M. Abdulrahman, N. Clarke, M. Brown, T. Kishida, M. Yao, F. Latif, E.R. Maher, Identification of candidate tumour suppressor genes frequently methylated in renal cell carcinoma. Oncogene 29(14), 2104–2117 (2010)
I.J. van Vlodrop, M.M. Baldewijns, K.M. Smits, L.J. Schouten, L. van Neste, W. van Criekinge, H. van Poppel, E. Lerut, K.E. Schuebel, N. Ahuja et al., Prognostic significance of Gremlin1 (GREM1) promoter CpG island hypermethylation in clear cell renal cell carcinoma. Am. J. Pathol. 176(2), 575–584 (2010)
Z. Li, X. Guo, Y. Wu, S. Li, J. Yan, L. Peng, Z. Xiao, S. Wang, Z. Deng, L. Dai et al., Methylation profiling of 48 candidate genes in tumor and matched normal tissues from breast cancer patients. Breast Cancer Res. Treat. 149(3), 767–779 (2015)
H. Kobayashi, K.A. Gieniec, J.A. Wright, T. Wang, N. Asai, Y. Mizutani, T. Lida, R. Ando, N. Suzuki, T.R.M. Lannagan et al., The balance of stromal BMP signaling mediated by GREM1 and ISLR drives colorectal carcinogenesis. Gastroenterology 160(4), 1224-1239.e1230 (2021)
T.C.M. Zuiverloon, F.C. de Jong, J.C. Costello, D. Theodorescu, Systematic review: characteristics and preclinical uses of bladder cancer cell lines. Bladder Cancer 4(2), 169–183 (2018)
C.F. Li, W.R. Wu, T.C. Chan, Y.H. Wang, L.R. Chen, W.J. Wu, B.W. Yeh, S.S Liang, Y.L. Shiue: transmembrane and coiled-coil domain 1 impairs the AKT signaling pathway in urinary bladder urothelial carcinoma: a characterization of a tumor suppressor. Clin. Cancer. Res. 23(24), 7650–7663 (2017)
T.C. Chan, W.J. Wu, W.M. Li, M.S. Shiao, Y.L. Shiue, C.F. Li, SLC14A1 prevents oncometabolite accumulation and recruits HDAC1 to transrepress oncometabolite genes in urothelial carcinoma. Theranostics 10(25), 11775–11793 (2020)
L.C. Li, R. Dahiya, MethPrimer: designing primers for methylation PCRs. Bioinformatics 18(11), 1427–1431 (2002)
W.R. Wu, J.T. Lin, C.T. Pan, T.C. Chan, C.L. Liu, W.J. Wu, J.J. Sheu, B.W. Yeh, S.K. Huang, J.Y. Jhung et al., Amplification-driven BCL6-suppressed cytostasis is mediated by transrepression of FOXO3 and post-translational modifications of FOXO3 in urinary bladder urothelial carcinoma. Theranostics 10(2), 707–724 (2020)
T. Wu, E. Hu, S. Xu, M. Chen, P. Guo, Z. Dai, T. Feng, L. Zhou, W. Tang, L. Zhan et al., clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb) 2(3), 100141 (2021)
A. Subramanian, P. Tamayo, V.K. Mootha, S. Mukherjee, B.L. Ebert, M.A. Gillette, A. Paulovich, S.L. Pomeroy, T.R. Golub, E.S. Lander et al., Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102(43), 15545–15550 (2005)
D.S. Chandrashekar, B. Bashel, S.A.H. Balasubramanya, C.J. Creighton, I. Ponce-Rodriguez, B. Chakravarthi, S. Varambally, UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19(8), 649–658 (2017)
J. Gao, B.A. Aksoy, U. Dogrusoz, G. Dresdner, B. Gross, S.O. Sumer, Y. Sun, A. Jacobsen, R. Sinha, E. Larsson et al., Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6(269), pl1 (2013)
A.G. Robertson, J. Kim, H. Al-Ahmadie, J. Bellmunt, G. Guo, A.D. Cherniack, T. Hinoue, P.W. Laird, K.A. Hoadley, R. Akbani et al., Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 171(3), 540-556.e525 (2017)
J. Xu, S. Lamouille, R. Derynck, TGF-beta-induced epithelial to mesenchymal transition. Cell. Res. 19(2), 156–172 (2009)
A. Liberzon, C. Birger, H. Thorvaldsdóttir, M. Ghandi, J.P. Mesirov, P. Tamayo, The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1(6), 417–425 (2015)
S. Lamouille, J. Xu, R. Derynck, Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15(3), 178–196 (2014)
P. Meylan, R. Dreos, G. Ambrosini, R. Groux, P. Bucher, EPD in 2020: enhanced data visualization and extension to ncRNA promoters. Nucleic Acids Res. 48(D1), D65-d69 (2020)
R.J. Wordinger, G. Zode, A.F. Clark, Focus on molecules: gremlin. Exp. Eye Res. 87(2), 78–79 (2008)
C. Zardecki, S. Dutta, D.S. Goodsell, R. Lowe, M. Voigt, S.K. Burley, PDB-101: educational resources supporting molecular explorations through biology and medicine. Protein Sci. 31(1), 129–140 (2022)
J. Mistry, S. Chuguransky, L. Williams, M. Qureshi, G.A. Salazar, E.L.L. Sonnhammer, S.C.E. Tosatto, L. Paladin, S. Raj, L.J. Richardson et al., Pfam: the protein families database in 2021. Nucleic Acids Res. 49(D1), D412-d419 (2021)
B. Jung, J.J. Staudacher, D. Beauchamp, Transforming growth factor β superfamily signaling in development of colorectal cancer. Gastroenterology 152(1), 36–52 (2017)
V.G. Martínez, C. Rubio, M. Martínez-Fernández, C. Segovia, F. López-Calderón, M.I. Garín, A. Teijeira, E. Munera-Maravilla, A. Varas, R. Sacedón et al., BMP4 induces M2 macrophage polarization and favors tumor progression in bladder cancer. Clin. Cancer Res. 23(23), 7388–7399 (2017)
L.M. Wakefield, C.S. Hill, Beyond TGFβ: roles of other TGFβ superfamily members in cancer. Nat. Rev. Cancer 13(5), 328–341 (2013)
M.K. Khokha, D. Hsu, L.J. Brunet, M.S. Dionne, R.M. Harland, Gremlin is the BMP antagonist required for maintenance of Shh and Fgf signals during limb patterning. Nat. Genet. 34(3), 303–307 (2003)
O. Korchynskyi, P. ten Dijke, Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 277(7), 4883–4891 (2002)
J. Perk, A. Iavarone, R. Benezra, Id family of helix-loop-helix proteins in cancer. Nat. Rev. Cancer 5(8), 603–614 (2005)
Z. Zhao, Z. Bo, W. Gong, Y. Guo, Inhibitor of differentiation 1 (Id1) in cancer and cancer therapy. Int. J. Med. Sci. 17(8), 995–1005 (2020)
B. Schmierer, C.S. Hill, TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat. Rev. Mol. Cell Biol. 8(12), 970–982 (2007)
A.P. Hinck, T.D. Mueller, T.A. Springer. Structural biology and evolution of the TGF-β family. Cold Spring Harb Perspect. Biol. 8(12), a022103 (2016)
L. Lan, T. Evan, H. Li, A. Hussain, E.J. Ruiz, M. Zaw Thin, R.M.M. Ferreira, H. Ps, E.M. Riising, Y. Zen et al., GREM1 is required to maintain cellular heterogeneity in pancreatic cancer. Nature 607(7917), 163–168 (2022)
D. Brenner, H. Blaser, T.W. Mak, Regulation of tumour necrosis factor signalling: live or let die. Nat. Rev. Immunol. 15(6), 362–374 (2015)
A. Yaron, A. Hatzubai, M. Davis, I. Lavon, S. Amit, A.M. Manning, J.S. Andersen, M. Mann, F. Mercurio, Y. Ben-Neriah, Identification of the receptor component of the IkappaBalpha-ubiquitin ligase. Nature 396(6711), 590–594 (1998)
P.A. Baeuerle, D. Baltimore, NF-kappa B: ten years after. Cell 87(1), 13–20 (1996)
T. Liu, L. Zhang, D. Joo, S.C. Sun, NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2, 17023- (2017)
M. Karin, Y. Ben-Neriah, Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu. Rev. Immunol. 18, 621–663 (2000)
C. Reily, T.J. Stewart, M.B. Renfrow, J. Novak, Glycosylation in health and disease. Nat. Rev. Nephrol. 15(6), 346–366 (2019)
N. Mitra, S. Sinha, T.N. Ramya, A. Surolia, N-linked oligosaccharides as outfitters for glycoprotein folding, form and function. Trends Biochem. Sci. 31(3), 156–163 (2006)
J.W. Lowery, J.M. Amich, A. Andonian, V. Rosen, N-linked glycosylation of the bone morphogenetic protein receptor type 2 (BMPR2) enhances ligand binding. Cell. Mol. Life Sci. 71(16), 3165–3172 (2014)
J.T. Stull, K.E. Kamm, R. Vandenboom, Myosin light chain kinase and the role of myosin light chain phosphorylation in skeletal muscle. Arch. Biochem. Biophys. 510(2), 120–128 (2011)
J.H. Wang, L. Zhang, S.T. Huang, J. Xu, Y. Zhou, X.J. Yu, R.Z. Luo, Z.S. Wen, W.H. Jia, M. Zheng, Expression and prognostic significance of MYL9 in esophageal squamous cell carcinoma. PLoS ONE 12(4), e0175280 (2017)
K. Matsushita, S. Kobayashi, H. Akita, M. Konno, A. Asai, T. Noda, Y. Iwagami, T. Asaoka, K. Gotoh, M. Mori, et al. Clinicopathological significance of MYL9 expression in pancreatic ductal adenocarcinoma. Cancer Rep. (Hoboken) 5(10), e1582 (2022)
Y. Deng, L. Liu, W. Feng, Z. Lin, Y. Ning, X. Luo, High expression of MYL9 indicates poor clinical prognosis of epithelial ovarian cancer. Recent Pat. Anticancer Drug Discov. 16(4), 533–539 (2021)
B.S. Kruthika, H. Sugur, K. Nandaki, A. Arimappamagan, K. Paturu, V. Santosh, Expression pattern and prognostic significance of myosin light chain 9 (MYL9): a novel biomarker in glioblastoma. J. Clin. Pathol. 72(10), 677–681 (2019)
Acknowledgements
We sincerely appreciate the technical assistance from Ms. SZ Dehghanian and Dr. TJ Chen.
Funding
The National Science and Technology Council (MOST-107–2314-B-110-MY3), NSYSU-KMU Joint Research Project (#NSYSUKMU 109-I008), Kaohsiung Armed Forces General Hospital (KAFGH-A-108021 & -109038), and Kaohsiung Medical University Research Center (KMU-TC111A02-0) supported this research.
Author information
Authors and Affiliations
Contributions
CFL, WJW, YLS conceived the concepts; CFL, CTP, YLS designed the experiments; TCC, CTP, HYH, PV, RJW, BWY, LRC, and MSS performed the experiments; CTP and CFL analyzed the data. CFL, CTP and YLS wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Human data and samples were obtained with informed consent, and the use was approved (IRB10207-001) by the Institutional Review Board of the Chi Mei Medical Center, Tainan, Taiwan. Animal treatments (#10626) were performed according to the Institutional Animal Care and Use Committee of National Sun Yat-sen University (NSYSU) Protocol, and NSYSU approved all protocols. This study does not contain any studies involving human participants performed by any of the authors.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chan, TC., Pan, CT., Hsieh, HY. et al. The autocrine glycosylated-GREM1 interacts with TGFB1 to suppress TGFβ/BMP/SMAD-mediated EMT partially by inhibiting MYL9 transactivation in urinary carcinoma. Cell Oncol. 46, 933–951 (2023). https://doi.org/10.1007/s13402-023-00788-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-023-00788-8