Abstract
Purpose
Glioblastoma (GBM) is the most common and deadly brain tumor. We aimed to reveal potential prognostic GBM marker genes, elaborate their functions, and build an effective a prognostic model for GBM patients.
Methods
Through data mining of The Cancer Genome Atlas (TCGA) and the Chinese Glioma Genome Atlas (CGGA), we screened for significantly differentially expressed genes (DEGs) to calculate risk scores for individual patients. Published data of somatic mutation and copy number variation profiles were analyzed for distinct genomic alterations associated with risk scores. In addition, single-cell sequencing was used to explore the biological functions of the identified prognostic marker genes. By combining risk scores and other clinical features, we built a comprehensive prognostic GBM model.
Results
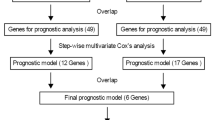
Seven DEGs (CLEC5A, HOXC6, HOXA5, CCL2, GPRASP1, BSCL2 and PTX3) were identified as being prognostic for GBM. Expression of these genes was confirmed in different GBM cell lines using real-time PCR. Risk scores calculated from the seven DEGs revealed prognostic value irrespective of other clinical factors, including IDH mutation status, and were negatively correlated with TP53 expression. The prognostic genes were found to be associated with tumor proliferation and progression based on pseudo-time analysis in neoplastic cells. A final prognostic model was developed and validated with a good performance, especially in geriatric GBM patients.
Conclusions
Using genetic profiles, age, IDH mutation status, and chemotherapy and radiotherapy, we constructed a comprehensive prognostic model for GBM patients. The model has a good performance, especially in geriatric GBM patients.








Similar content being viewed by others
Data availability
The datasets generated and analyzed during the current study are available in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/), TCGA data source (https://xena.ucsc.edu) and CGGA data portal (http://www.cgga.org.cn).
Abbreviations
- GBM:
-
Glioblastoma
- TCGA:
-
The Cancer Genome Atlas
- CGGA:
-
Chinese Glioma Genome Atlas
- DEGs:
-
differentially expressed genes
- GSVA:
-
geneset variation analysis
- WHO:
-
World Health Organization
- OS:
-
overall surviva
- ROC:
-
receiver operating characteristic
- AUC:
-
area under the curve
- CNV:
-
copy number variation
- GO:
-
gene ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- OPC:
-
oligodendrocyte precursor cell
- BP:
-
biological process
- MF:
-
molecular function
- NGS:
-
next-generation sequencing
References
Y. Yan, Z. Xu, S. Dai, L. Qian, L. Sun, Z. Gong, Targeting autophagy to sensitive glioma to temozolomide treatment. J. Exp. Clin. Cancer Res. 35, 23 (2016). https://doi.org/10.1186/s13046-016-0303-5
T. Jiang, Y. Mao, W. Ma, Q. Mao, Y. You, X. Yang, C. Jiang, C. Kang, X. Li, L. Chen, X. Qiu, W. Wang, W. Li, Y. Yao, S. Li, S. Li, A. Wu, K. Sai, H. Bai, G. Li, B. Chen, K. Yao, X. Wei, X. Liu, Z. Zhang, Y. Dai, S. Lv, L. Wang, Z. Lin, J. Dong, G. Xu, X. Ma, J. Cai, W. Zhang, H. Wang, L. Chen, C. Zhang, P. Yang, W. Yan, Z. Liu, H. Hu, J. Chen, Y. Liu, Y. Yang, Z. Wang, Z. Wang, Y. Wang, G. You, L. Han, Z. Bao, Y. Liu, Y. Wang, X. Fan, S. Liu, X. Liu, Y. Wang, Q. Wang, G. Chinese Glioma Cooperative, CGCG clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 375, 263–273 (2016). https://doi.org/10.1016/j.canlet.2016.01.024
H. Zhang, R. Wang, Y. Yu, J. Liu, T. Luo, F. Fan, Glioblastoma treatment modalities besides surgery. J. Cancer 10, 4793–4806 (2019). https://doi.org/10.7150/jca.32475
X. Chen, L. Zhang, I.Y. Zhang, J. Liang, H. Wang, M. Ouyang, S. Wu, A.C.C. da Fonseca, L. Weng, Y. Yamamoto, H. Yamamoto, R. Natarajan, B. Badie, RAGE expression in tumor-associated macrophages promotes angiogenesis in glioma. Cancer Res. 74, 7285–7297 (2014). https://doi.org/10.1158/0008-5472.CAN-14-1240
K. Yang, Y. Liu, Z. Liu, J. Liu, X. Liu, X. Chen, C. Li, Y. Zeng, p38gamma overexpression in gliomas and its role in proliferation and apoptosis. Sci. Rep. 3, 2089 (2013). https://doi.org/10.1038/srep02089
J. Huang, F. Liu, Z. Liu, H. Tang, H. Wu, Q. Gong, J. Chen, Immune checkpoint in glioblastoma: promising and challenging. Front. Pharmacol. 8, 242 (2017). https://doi.org/10.3389/fphar.2017.00242
W.S. Wen, S.L. Hu, Z. Ai, L. Mou, J.M. Lu, S. Li, Methylated of genes behaving as potential biomarkers in evaluating malignant degree of glioblastoma. J. Cell. Physiol. 232, 3622–3630 (2017). https://doi.org/10.1002/jcp.25831
M.M. Binabaj, A. Bahrami, S. ShahidSales, M. Joodi, M. Joudi Mashhad, S.M. Hassanian, K. Anvari, A. Avan, The prognostic value of MGMT promoter methylation in glioblastoma: A meta-analysis of clinical trials. J. Cell. Physiol. 233, 378–386 (2018). https://doi.org/10.1002/jcp.25896
H. Zhang, Y. Zhou, B. Cui, Z. Liu, H. Shen, Novel insights into astrocyte-mediated signaling of proliferation, invasion and tumor immune microenvironment in glioblastoma. Biomed. Pharmacother. 126, 110086 (2020). https://doi.org/10.1016/j.biopha.2020.110086
Z. Wang, H. Zhang, Q. Cheng, PDIA4, The basic characteristics, functions and its potential connection with cancer. Biomed. Pharmacother. 122, 109688 (2020). https://doi.org/10.1016/j.biopha.2019.109688
H. Zhang, B. Cui, Y. Zhou, X. Wang, W. Wu, Z. Wang, Z. Dai, Q. Cheng, K. Yang, B2M overexpression correlates with malignancy and immune signatures in human gliomas. Sci. Rep. 11, 5045 (2021). https://doi.org/10.1038/s41598-021-84465-6
H. Zhang, J. He, Z. Dai, Z. Wang, X. Liang, F. He, Z. Xia, S. Feng, H. Cao, L. Zhang, Q. Cheng, PDIA5 is Correlated with immune infiltration and predicts poor prognosis in gliomas. Front. Immunol. 12, 628966 (2021). https://doi.org/10.3389/fimmu.2021.628966
H. Zhang, Z. Chen, Z. Wang, Z. Dai, Z. Hu, X. Zhang, M. Hu, Z. Liu, Q. Cheng, Correlation between APOBEC3B expression and clinical characterization in lower-grade gliomas. Front. Oncol. 11, 625838 (2021). https://doi.org/10.3389/fonc.2021.625838
N. Zhang, Z. Dai, W. Wu, Z. Wang, H. Cao, Y. Zhang, Z. Wang, H. Zhang, Q. Cheng, The predictive value of monocytes in immune microenvironment and prognosis of glioma patients based on machine learning. Front. Immunol. 12, 656541 (2021). https://doi.org/10.3389/fimmu.2021.656541
L. Gautier, L. Cope, B.M. Bolstad, R.A. Irizarry, affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315 (2004). https://doi.org/10.1093/bioinformatics/btg405
J.T. Leek, W.E. Johnson, H.S. Parker, A.E. Jaffe, J.D. Storey, The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012). https://doi.org/10.1093/bioinformatics/bts034
M.E. Ritchie, B. Phipson, D. Wu, Y. Hu, C.W. Law, W. Shi, G.K. Smyth, limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). https://doi.org/10.1093/nar/gkv007
J.J. Goeman, L1 penalized estimation in the Cox proportional hazards model. Biom. J. 52, 70–84 (2010). https://doi.org/10.1002/bimj.200900028
S. Hanzelmann, R. Castelo, J. Guinney, GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 14, 7 (2013). https://doi.org/10.1186/1471-2105-14-7
P. Geeleher, N. Cox, R.S. Huang, pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One 9, e107468 (2014). https://doi.org/10.1371/journal.pone.0107468
T. Stuart, A. Butler, P. Hoffman, C. Hafemeister, E. Papalexi, W.M. Mauck 3rd, Y. Hao, M. Stoeckius, P. Smibert, R. Satija, Comprehensive integration of single-cell data. Cell 177, 1888–1902 e1821 (2019). https://doi.org/10.1016/j.cell.2019.05.031
R. Wang, R. Sharma, X. Shen, A.M. Laughney, K. Funato, P.J. Clark, M. Shpokayte, P. Morgenstern, M. Navare, Y. Xu, S. Harbi, I. Masilionis, G. Nanjangud, Y. Yang, G. Duran-Rehbein, M. Hemberg, D. Pe’er, V. Tabar, Adult human glioblastomas harbor radial glia-like cells. Stem Cell Rep. 14, 338–350 (2020). https://doi.org/10.1016/j.stemcr.2020.01.007
M.A. Tosches, T.M. Yamawaki, R.K. Naumann, A.A. Jacobi, G. Tushev, G. Laurent, Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science 360, 881–888 (2018). https://doi.org/10.1126/science.aar4237
X. Han, Z. Zhou, L. Fei, H. Sun, R. Wang, Y. Chen, H. Chen, J. Wang, H. Tang, W. Ge, Y. Zhou, F. Ye, M. Jiang, J. Wu, Y. Xiao, X. Jia, T. Zhang, X. Ma, Q. Zhang, X. Bai, S. Lai, C. Yu, L. Zhu, R. Lin, Y. Gao, M. Wang, Y. Wu, J. Zhang, R. Zhan, S. Zhu, H. Hu, C. Wang, M. Chen, H. Huang, T. Liang, J. Chen, W. Wang, D. Zhang, G. Guo, Construction of a human cell landscape at single-cell level. Nature. 581, 303–309 (2020). https://doi.org/10.1038/s41586-020-2157-4
B. Pang, J. Xu, J. Hu, F. Guo, L. Wan, M. Cheng, L. Pang, Single-cell RNA-seq reveals the invasive trajectory and molecular cascades underlying glioblastoma progression. Mol. Oncol. 13, 2588–2603 (2019). https://doi.org/10.1002/1878-0261.12569
X. Robin, N. Turck, A. Hainard, N. Tiberti, F. Lisacek, J.C. Sanchez, M. Muller, pROC: an open-source package for R and S + to analyze and compare ROC curves. BMC Bioinf. 12, 77 (2011). https://doi.org/10.1186/1471-2105-12-77
M. Zhang, X. Wang, X. Chen, Q. Zhang, J. Hong, Novel immune-related gene signature for risk stratification and prognosis of survival in lower-grade glioma. Front. Genet. 11, 363 (2020). https://doi.org/10.3389/fgene.2020.00363
M. Cao, J. Cai, Y. Yuan, Y. Shi, H. Wu, Q. Liu, Y. Yao, L. Chen, W. Dang, X. Zhang, J. Xiao, K. Yang, Z. He, X. Yao, Y. Cui, X. Zhang, X. Bian, A four-gene signature-derived risk score for glioblastoma: prospects for prognostic and response predictive analyses. Cancer Biol. Med. 16, 595–605 (2019). https://doi.org/10.20892/j.issn.2095-3941.2018.0277
H.W. Fan, Q. Ni, Y.N. Fan, Z.X. Ma, Y.B. Li, C-type lectin domain family 5, member A (CLEC5A, MDL-1) promotes brain glioblastoma tumorigenesis by regulating PI3K/Akt signalling. Cell Prolif. e12584 (2019). https://doi.org/10.1111/cpr.12584
T.F. Yan, M.J. Wu, B. Xiao, Q. Hu, Y.H. Fan, X.G. Zhu, Knockdown of HOXC6 inhibits glioma cell proliferation and induces cell cycle arrest by targeting WIF-1 in vitro and vivo. Pathol. Res. Pract. 214, 1818–1824 (2018). https://doi.org/10.1016/j.prp.2018.09.001
G. Arderiu, I. Cuevas, A. Chen, M. Carrio, L. East, N.J. Boudreau, HoxA5 stabilizes adherens junctions via increased Akt1. Cell Adhes. Migr. 1, 185–195 (2007). https://doi.org/10.4161/cam.1.4.5448
P.J. Cimino, Y. Kim, H.J. Wu, J. Alexander, H.G. Wirsching, F. Szulzewsky, K. Pitter, T. Ozawa, J. Wang, J. Vazquez, S. Arora, R. Rabadan, R. Levine, F. Michor, E.C. Holland, Increased HOXA5 expression provides a selective advantage for gain of whole chromosome 7 in IDH wild-type glioblastoma. Genes Dev. 32, 512–523 (2018). https://doi.org/10.1101/gad.312157.118
B. Lu, Y. Zhou, Z. Su, A. Yan, P. Ding, Effect of CCL2 siRNA on proliferation and apoptosis in the U251 human glioma cell line. Mol. Med. Rep. 16, 3387–3394 (2017). https://doi.org/10.3892/mmr.2017.6995
A.L. Chang, J. Miska, D.A. Wainwright, M. Dey, C.V. Rivetta, D. Yu, D. Kanojia, K.C. Pituch, J. Qiao, P. Pytel, Y. Han, M. Wu, L. Zhang, C.M. Horbinski, A.U. Ahmed, M.S. Lesniak, CCL2 produced by the glioma microenvironment is essential for the recruitment of regulatory T cells and myeloid-derived suppressor cells. Cancer Res. 76, 5671–5682 (2016). https://doi.org/10.1158/0008-5472.CAN-16-0144
E. Mori, J. Fujikura, M. Noguchi, K. Nakao, M. Matsubara, M. Sone, D. Taura, T. Kusakabe, K. Ebihara, T. Tanaka, K. Hosoda, K. Takahashi, I. Asaka, N. Inagaki, K. Nakao, Impaired adipogenic capacity in induced pluripotent stem cells from lipodystrophic patients with BSCL2 mutations. Metabolism 65, 543–556 (2016). https://doi.org/10.1016/j.metabol.2015.12.015
L. Liu, Q. Jiang, X. Wang, Y. Zhang, R.C. Lin, S.M. Lam, G. Shui, L. Zhou, P. Li, Y. Wang, X. Cui, M. Gao, L. Zhang, Y. Lv, G. Xu, G. Liu, D. Zhao, H. Yang, Adipose-specific knockout of SEIPIN/BSCL2 results in progressive lipodystrophy. Diabetes 63, 2320–2331 (2014). https://doi.org/10.2337/db13-0729
L. Wirestam, H. Enocsson, T. Skogh, M.L. Eloranta, L. Ronnblom, C. Sjowall, J. Wettero, Interferon-alpha coincides with suppressed levels of pentraxin-3 (PTX3) in systemic lupus erythematosus and regulates leucocyte PTX3 in vitro. Clin. Exp. Immunol. 189, 83–91 (2017). https://doi.org/10.1111/cei.12957
C. Garlanda, B. Bottazzi, A. Bastone, A. Mantovani, Pentraxins at the crossroads between innate immunity, inflammation, matrix deposition, and female fertility. Annu. Rev. Immunol. 23, 337–366 (2005). https://doi.org/10.1146/annurev.immunol.23.021704.115756
Q. Liu, X.Y. Wang, Y.Y. Qin, X.L. Yan, H.M. Chen, Q.D. Huang, J.K. Chen, J.M. Zheng, SPOCD1 promotes the proliferation and metastasis of glioma cells by up-regulating PTX3. Am. J. Cancer Res. 8, 624–635 (2018)
M. Locatelli, S. Ferrero, F. Martinelli Boneschi, L. Boiocchi, M. Zavanone, S. Maria Gaini, L. Bello, S. Valentino, E. Barbati, M. Nebuloni, A. Mantovani, C. Garlanda, The long pentraxin PTX3 as a correlate of cancer-related inflammation and prognosis of malignancy in gliomas. J. Neuroimmunol. 260, 99–106 (2013). https://doi.org/10.1016/j.jneuroim.2013.04.009
Z. Chen, X. Feng, C.J. Herting, V.A. Garcia, K. Nie, W.W. Pong, R. Rasmussen, B. Dwivedi, S. Seby, S.A. Wolf, D.H. Gutmann, D. Hambardzumyan, Cellular and molecular identity of tumor-associated macrophages in glioblastoma. Cancer Res. 77, 2266–2278 (2017). https://doi.org/10.1158/0008-5472.CAN-16-2310
S. Nakae, T. Kato, K. Murayama, H. Sasaki, M. Abe, M. Kumon, T. Kumai, K. Yamashiro, J. Inamasu, M. Hasegawa, H. Kurahashi, Y. Hirose, Remote intracranial recurrence of IDH mutant gliomas is associated with TP53 mutations and an 8q gain. Oncotarget 8, 84729–84742 (2017). https://doi.org/10.18632/oncotarget.20951
T. Yang, P. Mao, X. Chen, X. Niu, G. Xu, X. Bai, W. Xie, Inflammatory biomarkers in prognostic analysis for patients with glioma and the establishment of a nomogram. Oncol. Lett. 17, 2516–2522 (2019). https://doi.org/10.3892/ol.2018.9870
M. Lacroix, D. Abi-Said, D.R. Fourney, Z.L. Gokaslan, W. Shi, F. DeMonte, F.F. Lang, I.E. McCutcheon, S.J. Hassenbusch, E. Holland, K. Hess, C. Michael, D. Miller, R. Sawaya, A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extent of resection, and survival. J. Neurosurg. 95, 190–198 (2001). https://doi.org/10.3171/jns.2001.95.2.0190
Q.T. Ostrom, H. Gittleman, J. Fulop, M. Liu, R. Blanda, C. Kromer, Y. Wolinsky, C. Kruchko, J.S. Barnholtz-Sloan, CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro Oncol. 17(4), iv1–iv62 (2015). https://doi.org/10.1093/neuonc/nov189
Acknowledgements
We would like to thank Prof. Bing Jiang and Prof. Zhixiong Liu for their assistance in this research, and Dr. Chris Lou for bioinformatics support.
Funding
This work was supported by the Hunan provincial health and Health Committee Foundation of China (C2019186), the China Postdoctoral Science Foundation (NO.2018M633002), the Hunan Provincial Natural Science Foundation of China (NO.2018JJ3838) and the Science and Technology Department of Hunan Province (NO.2015SK2032-2).
Author information
Authors and Affiliations
Contributions
Quan Cheng has made substantial contributions to the conception and design of the study, the acquisition of data and the analysis and interpretation of data. Fan Fan, Hao Zhang, Nan Zhang and Ziyu Dai have been involved in drafting the manuscript and revising it. Yakun Zhang, Zhiwei Xia, Kui Yang, Fengqin Ding and Shui Hu provided technical assistance. Hui Cao and Yong Guo provided writing assistance.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Fan, F., Zhang, H., Dai, Z. et al. A comprehensive prognostic signature for glioblastoma patients based on transcriptomics and single cell sequencing. Cell Oncol. 44, 917–935 (2021). https://doi.org/10.1007/s13402-021-00612-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-021-00612-1