Abstract
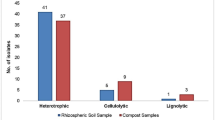
An endeavour is underway to isolate native cellulose-degrading bacteria from different sources in order to have them used in composting of organic waste. On the basis of cellulase production and growth in different environmental circumstances, five isolates were selected. The 16S rRNA genetic sequence of five isolates was submitted to the National Center for Biotechnology Information (NCBI), which assigned accession numbers ON150745, ON178665, ON725042, ON479186 and ON142173 for CBD4, CBG3, CBG2, CBC9 and CBG4 in accordance. The growth of CBD4, CBG2, CBG3 and CBG4 reached the stationary phase at 24 h and lasted up to 78 h, but in case of CBC9 stationary phase of growth lies between 32 and 50 h of incubation which was less than other bacterial strains. The strains’ optimum CMCase production conditions were 5% inoculum, a pH of 7.5, the temperature of 35° C for CBD4 and CBC9; 4% inoculum, a pH of 6.5 and temperature of 35° C for CBG2, CBG3; and 3% inoculum with same pH and temperature for CBG4. The CMCase production at respective optimum conditions was highest in the case of CBG2 strain. The CMCase reaction for all the strains was highest at 50 °C and decreased in higher or lower temperatures. The reaction of cellulase produced by strain CBG2 at optimum pH and temperature was highest. In this study, the cellulolytic potential of the CBC9 strain (B. xiamenensis) was examined for the first time.












Similar content being viewed by others
References
Hemati A, Nazari M, Lajayer BA, Smith DL, Astatkie T (2022) Lignocellulosics in plant cell wall and their potential biological degradation. Folia Microbiol 67(5):671–681. https://doi.org/10.1007/s12223-022-00974-5
Menshawy MN, Abdel-Hamid AM, Mohamed SK, Mo’men H, (2022) Isolation and molecular identification of cellulose/hemicellulose degrading bacteria from agricultural compost and determination of their hydrolytic potential. S Afr J Bot 149:617–621
Yang W, Meng F, Peng J, Han P, Fang F, Ma L, Cao B (2014) Isolation and identification of a cellulolytic bacterium from the Tibetan pig’s intestine and investigation of its cellulase production. Electron J Biotechnol 17(6):262–267
Pandit L, Sethi D, Pattanayak SK, Nayak Y (2020) Bioconversion of lignocellulosic organic wastes into nutrient rich vermicompost by Eudrilus eugeniae. Bioresour Technol Rep 12:100580
Patra RK, Behera D, Mohapatra KK, Sethi D, Mandal M, Patra AK, Balasubramani R (2022) Juxtaposing the quality of compost and vermicompost produced from organic waste amended with cow dung. Environ Res 214:114119. https://doi.org/10.1016/j.envres.2022.114119
Zhang T, Wu X, Shaheen SM, Abdelrahman H, Ali EF, Bolan NS, Ok YS, Li G, Tsang DCW, Rinklebe J (2022) Improving the humification and phosphorus flow during swine manure composting: a trial for enhancing the beneficial applications of hazardous biowastes. J Hazard Mater 425:127906. https://doi.org/10.1016/j.jhazmat.2021.127906
Vodovnik M, Marinsek-Logar R (2010) Cellulosomes - promising supramolecular machines of anaerobic cellulolytic microorganisms. Acta Chim Slov 57(4):767–774
Delangiz N, Aliyar S, Pashapoor N, Nobaharan K, Lajayer BA, Rodríguez-Coutoe S (2022) Can polymer-degrading microorganisms solve the bottleneck of plastics’ environmental challenges? Chemosphere 294:133709. https://doi.org/10.1016/j.chemosphere.2022.133709
Yu H, Zeng G, Huang H, Xi X, Wang R, Huang D, Huang G, Li J (2007) Microbial community succession and lignocellulose degradation during agricultural waste composting. Biodegradation 18(6):793–802
Maki M, Leung KT, Qin W (2009) The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. Int J Biol Sci 5(5):500–516
Doi RH (2008) Cellulases of mesophilic microorganisms: cellulosome and noncellulosome producers. Ann N Y Acad Sci 1125(1):267–279
Dar MA, Shaikh AA, Pawar KD, Pandit RS (2018) Exploring the gut of Helicoverpa armigera for cellulose degrading bacteria and evaluation of a potential strain for lignocellulosic biomass deconstruction. Process Biochem 73:142–153
Prasad RK, Chatterjee S, Mazumder PB, Sharma S, Datta S, Vairale MG, Dwivedi SK (2019) Study on cellulase (Β-1, 4-endoglucanase) activity of gut bacteria of Sitophilus oryzae in cellulosic waste biodegradation. Bioresour Technol Rep 7:100274
Bhuyan PM, Sandilya SP, Nath PK, Gandotra S, Subramanian S, Kardong D, Gogoi DK (2018) Optimization and characterization of extracellular cellulase produced by Bacillus pumilus MGB05 isolated from midgut of muga silkworm (Antheraea assamensis Helfer). J Asia Pac Entomol 21(4):1171–1181
Saha S, Khatun F, Nahiduzzaman M, Mahmud MP, Rahman MM, Yasmin S (2022) A greener way of lactic acid production from cellulose-rich waste materials utilising cellulolytic bacteria population from insect gut. Bioresour Technol Rep 18:101021
Mmango-Kaseke Z, Okaiyeto K, Nwodo UU, Mabinya LV, Okoh AI (2016) Optimization of cellulase and xylanase production by Micrococcus species under submerged fermentation. Sustainability 8(11):1168
Hemati A, Aliasgharzad N, Khakvar R, Khoshmanzar E, Lajayer BA, van Hullebusch ED (2021) Role of lignin and thermophilic lignocellulolytic bacteria in the evolution of humification indices and enzymatic activities during compost production. Waste Manage 119:122–134. https://doi.org/10.1016/j.wasman.2020.09.042
Yang Y, Dou Y, Wang B, Xue Z, Wang Y, An S, Chang SX (2022) Deciphering factors driving soil microbial life-history strategies in restored grassland. iMeta 2022: 66. https://doi.org/10.1002/imt2.66.
Balasubramanian N, Simões N (2014) Bacillus pumilus S124A carboxymethyl cellulase; a thermo stable enzyme with a wide substrate spectrum utility. Int J Biol Macromol 67:132–139
Li F, Xie Y, Gao X, Shan M, Sun C, Niu YD, Shan A (2020) Screening of cellulose degradation bacteria from Min pigs and optimization of its cellulase production. Electron J Biotechnol 48:29–35
Gupta P, Samant K, Sahu A (2012) Isolation of cellulose-degrading bacteria and determination of their cellulolytic potential. Int J Microbiol 2012: 578925. doi:https://doi.org/10.1155/2012/578925.
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31(3):426–428. https://doi.org/10.1021/ac60147a030
Verma H, Patra RK, Sethi D, Pattanayak SK (2022) Isolation and characterization of native Rhizobium from root nodules of Raikia french bean growing area of Odisha. Indian J Biochem Biophys 59:918–926. https://doi.org/10.56042/ijbb.v59i9.61519
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547
Stecher G, Tamura K, Kumar S (2020) Molecular evolutionary genetics analysis (MEGA) for macOS. Mol Biol Evol 37:1237
R Core Team (2021) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.
Islam F, Roy N (2018) Screening, purification and characterization of cellulase from cellulase producing bacteria in molasses. BMC res Notes 11(1):1–6
Ferbiyanto A, Rusmana I, Raffiudin R (2015) Characterization and identification of cellulolytic bacteria from gut of worker Macrotermes gilvus. HAYATI J of Biosc 22:197–200
Subudhi S, Sethi D, Pattanayak SK (2020) Characterization of Rhizobium sp (SAR-5) isolated from root nodule of Acacia mangium L. Indian J Biochem Biophys 57:327–333
Ye M, Tang X, Yang R, Zhang H, Li F, Tao F, Li F, Wang Z (2018) Characteristics and application of a novel species of Bacillus: Bacillus velezensis. ACS Chem Biol 13(3):500–505
Lai Q, Liu Y, Shao Z (2014) Bacillus xiamenensis sp. nov., isolated from intestinal tract contents of a flathead mullet (Mugil cephalus). Antonie Leeuwenhoek 105:99–107
Peixoto SB, Florencia CO, Daniel JD, Adriano B (2011) Cellulase-producing Bacillus strains isolated from the intestine of Amazon basin fish. Aquac Res 42:887–891
Raza A, Bashir S, Tabassum R (2019) Evaluation of cellulases and xylanases production from Bacillus spp isolated from buffalo digestive system Kafkas. Univ Vet Fak Derg 25(1):39–46
Okaiyeto K, Nwodo UU, Okoli SA, Mabinya LV, Okoh AI (2016) Implications for public health demands alternatives to inorganic and synthetic flocculants: bioflocculants as important candidates. Microbiol Open 5(2):177–211
Azadian F, Badoei-Dalfard A, Namaki-Shoushtari A, Karami Z, Hassanshahian M (2017) Production and characterization of an acido-thermophilic, organic solvent stable cellulase from Bacillus sonorensis HSC7 by conversion of lignocellulosic wastes. J Genet Eng Biotechnol 15(1):187–196
Hussain AA, Abdel-Salam MS, Abo-Ghalia HH, Hegazy WK, Hafez SS (2017) Optimization and molecular identification of novel cellulose degrading bacteria isolated from Egyptian environment. J Genet Eng Biotechnol 15(1):77–85
Simões MLG, Tauk-Tornisielo MS, Tapia DMT (2009) Screening of culture condition for xylanase production by filamentous fungi. Afr J Biotechnol 8(22):6317–6326
Ire FS, Ezebuiro V, Ogugbue CJ (2016) Production of bioethanol by bacterial co-culture from agro-waste-impacted soil through simultaneous saccharification and co-fermentation of steam-exploded bagasse. Bioresour Bioprocess 3:26. https://doi.org/10.1186/s40643-016-0104-x
Sreena CP, Sebastian D (2018) Augmented cellulase production by Bacillus subtilis strain MU S1 using different statistical experimental designs. J Genet Eng Biotechnol 16:9–16
Liang YL, Zhang Z, Wu M, Wu Y, Feng JX (2014) Isolation, screening, and identification of cellulolytic bacteria from natural reserves in the subtropical region of China and optimization of cellulase production by Paenibacillus terrae ME27–1. Bio Med Res Int 2014:512497. https://doi.org/10.1155/2014/512497
Chantarasiri A (2015) Aquatic Bacillus cereus JD0404 isolated from the muddy sediments of mangrove swamps in Thailand and characterization of its cellulolytic activity. Egypt J Aquat Res 41(3):257–264
Acknowledgements
The infrastructure support of Odisha University of Agriculture and Technology is duly acknowledged.
Funding
The financial support of Odisha University of Agriculture and Technology is duly acknowledged.
Author information
Authors and Affiliations
Contributions
Kshitipati Padhan, Ranjan Kumar Patra: conceptualization, writing—original draft, review and editing; Akshaya Kumar Senapati: conceptualization, writing—review and editing; Debadatta Sethi: writing—original draft, writing—review and editing; Narayan Panda: writing—review and editing; Sanjib Kumar Sahoo: writing—review and editing; Sushanta Kumar Pattanayak: conceptualization, writing—review and editing.
Corresponding author
Ethics declarations
Ethical approval
No human/animal studies were involved in this study.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Padhan, K., Patra, R.K., Sethi, D. et al. Isolation, characterization and identification of cellulose-degrading bacteria for composting of agro-wastes. Biomass Conv. Bioref. (2023). https://doi.org/10.1007/s13399-023-04087-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13399-023-04087-y