Abstract
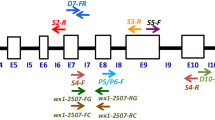
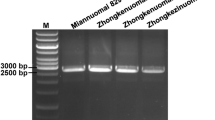
Waxy corn possessing high amylopectin is widely employed as an industrial product. Traditional corn contains ~ 70–75% amylopectin, whereas waxy corn with the mutant waxy1 (wx1) gene possesses ~ 95–100% amylopectin. Marker-assisted breeding can greatly hasten the transfer of the wx1 allele into normal corn. However, the available gene-based marker(s) for wx1 are not always polymorphic between recipient and donor parents, thereby causing a considerable delay in the molecular breeding program. Here, a 4800 bp sequence of the wx1 gene was analyzed among seven wild-type and seven mutant inbreds employing 16 overlapping primers. Three polymorphisms viz., 4 bp InDel (at position 2406 bp) in intron-7 and two SNPs (C to A at position 3325 bp in exon-10 and G to T at position 4310 bp in exon-13) differentiated the dominant (Wx1) and recessive (wx1) allele. Three breeder-friendly PCR markers (WxDel4, SNP3325_CT1, and SNP4310_GT2) specific to InDel and SNPs were developed. WxDel4 amplified 94 bp among mutant-type inbreds, while 90 bp was amplified among wild-type inbreds. SNP3325_CT1 and SNP4310_GT2 revealed the presence-absence polymorphisms with an amplification of 185 bp and 189 bp of amplicon, respectively. These newly developed markers showed 1:1 segregation in both BC1F1 and BC2F1 generations, while 1:2:1 segregation was observed in BC2F2. The recessive homozygotes (wx1wx1) of BC2F2 identified by the markers possessed significantly higher amylopectin (97.7%) compared to the original inbreds (Wx1Wx1: 72.7% amylopectin). This is the first report of novel wx1 gene-based markers. The information generated here would help in accelerating the development of waxy maize hybrids.





Similar content being viewed by others
References
Abhijith KP, Muthusamy V, Chhabra R, Dosad S, Bhatt V, Chand G, Jaiswal SK, Zunjare RU, Vasudev S, Yadava DK, Hossain F (2020) Development and validation of breeder-friendly gene-based markers for lpa1–1 and lpa2–1 genes conferring low phytic acid in maize kernel. 3 Biotech 10:121. https://doi.org/10.1007/s13205-020-2113-x
Bao JD, Yao JQ, Zhu JQ (2012) Identification of glutinous maize landraces and inbred lines with altered transcription of waxy gene. Mol Breed 30:1707–1714
Brimhall B, Sprague GF, Sass JE (1945) A new waxy allele in corn and its effect on the properties of the endosperm starch. J Agronomy 37:937–944
Chhabra R, Hossain F, Muthusamy V, Baveja A, Mehta BK, Zunjare RU (2019) Development and validation of breeder-friendly functional markers of sugary1 gene encoding starch-debranching enzyme affecting kernel sweetness in maize (Zea mays). Crop Pasture Sci 70(10):868–875. https://doi.org/10.1071/CP19298
Chhabra R, Hossain F, Muthusamy V, Baveja A, Mehta BK, Zunjare RU (2020) Development and validation of gene-based markers for shrunken2-Reference allele and their utilization in marker-assisted sweet corn (Zea mays Sachharata) breeding programme. Plant Breed :1–10. https://doi.org/10.1111/pbr.12872
Devi EL, Chhabra R, Jaiswal SK, Hossain F, Zunjare RU, Goswami R, Muthusamy V, Baveja A, Dosad S (2017) Microsatellite marker-based characterization of waxy maize inbreds for their utilization in hybrid breeding. 3 Biotech 7:316
Dong L, Qi X, Zhu J, Liu C, Zhang X, Cheng B, Mao L, Xie C (2019) Supersweet and waxy: meeting the diverse demands for specialty maize by genome editing. Plant Biotechnol J 17(10):1853–1855. https://doi.org/10.1111/pbi.13144
Flint-Garcia SA, Thornsberry JM, Buckler ES (2003) Structure of linkage disequilibrium in plants. Ann Rev Plant Biol 54:357–374. https://doi.org/10.1146/annurev.arplant.54.031902.134907
Gupta HS, Babu R, Agrawal PK, Mahajan V, Hossain F, Nepolean T (2013) Accelerated development of quality protein maize hybrid through marker-assisted introgression of opaque-2 allele. Plant Breed 132:77–82
Hossain F, Muthusamy V, Pandey N, Vishwakarma AK, Baveja A, Zunjare RU, Thirunavukkarasu N, Saha S, Manjaiah KM, Prasanna BM, Gupta HS (2018) Marker-assisted introgression of opaque2 allele for rapid conversion of elite hybrids into quality protein maize. J Genet 97:287–298
Hossain F, Chhabra R, Devi EL, Zunjare RU, Jaiswal SK, Muthusamy V (2019) Molecular analysis of mutant granule-bound starch synthase-I (waxy1) gene in diverse waxy maize inbreds. 3 Biotech 9:3. https://doi.org/10.1007/s13205-018-1530-6
Huang BQ, Tian ML, Zhang JJ, Huang YB (2010) Waxy locus and its mutant types in maize (Zea mays L). J Integr Agr 9:1–10
Klosgen RB, Gierl A, Schwarz-Sommer Z, Saedler H (1986) Molecular analysis of the waxy locus of Zea mays. Mol Genet Genom 203:237–244
Lin F, Zhou L, He B, Zhang X, Dai H, Qian Y, Ruan L, Zhao H (2019) QTL mapping for maize starch content and candidate gene prediction combined with co-expression network analysis. Theor Appl Genet 132(7):1931–1941
Liu J, Rong T, Li W (2007) Mutation loci and intragenic selection marker of the granule-bound starch synthase gene in waxy maize. Mol Breed 20:93–102. https://doi.org/10.1007/s11032-006-9074-6
Liu J, Huang S, Sun M (2012) An improved allele-specific PCR primer design method for SNP marker analysis and its application. Plant Methods 8:34. https://doi.org/10.1186/1746-4811-8-34
Liu H, Wang X, Wei B, Wang Y, Liu Y, Zhang J, Hu Y, Yu G, Li J, Xu Z, Huang Y (2016) Characterization of genome-wide variation in Four-row Wax, a waxy maize landrace with a reduced kernel row phenotype. Front Plant Sci 7:667. https://doi.org/10.3389/fpls.2016.00667
Macdonald FD, Preiss J (1985) Partial purification and characterization of granule-bound starch synthases from normal and waxy maize. J Plant Physiol 78:849–852
Marillonnet S, Wessler SR (1997) Retrotransposon insertion into the maize waxy gene results in tissue-specific RNA processing. Plant Cell 9:967–978
Mason-Gamer RJ, Well CF, Kellogg EA (1998) Granule-bound starch synthase: structure, function, and phylogenetic utility. Mol Biol Evol 15:1658–1673
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326. https://doi.org/10.1093/nar/8.19.4321
Muthusamy V, Hossain F, Thirunavukkarasu N, Choudhary M, Saha S, Bhat JS, Prasanna BM, Gupta HS (2014) Development of β-carotene rich maize hybrids through marker-assisted introgression of β-carotene hydroxylase allele. PLoS ONE 9:e113583
Nelson OE, Rines HW (1962) The enzymatic deficiency in the waxy mutant of maize. Biochem Biophy Res Commun 9:297–300
Okagaki RJ, Neuffer MG, Wessler SR (1991) A deletion common to two independently derived waxy mutations of maize. Genetics 128:425–431
Qi X, Dong L, Liu C, Mao L, Liu F, Zhang X, Cheng B, Xie C (2018) Systematic identification of endogenous RNA polymerase III promoters for efficient RNA guide-based genome editing technologies in maize. Crop J 6:314–320
Qi X, Wu H, Jiang H, Zhu J, Huang C, Zhang X, Liu C, Cheng B (2020) Conversion of a normal maize hybrid into a waxy version using in vivo CRISPR/Cas9 targeted mutation activity. Crop J. https://doi.org/10.1016/j.cj.2020.01.006
Reddappa SB, Chhabra R, Talukder ZA, Muthusamy V, Zunjare RU, Hossain F (2022) Development and validation of rapid and cost-effective protocol for estimation of amylose and amylopectin in maize kernels. 3 Biotech 12:62. https://doi.org/10.1007/s13205-022-03128-z
Sarika K, Hossain F, Muthusamy V, Zunjare RU, Baveja A, Goswami R, Bhat JS, Saha S, Gupta HS (2018) Marker-assisted pyramiding of opaque2 and novel opaque16 genes for further enrichment of lysine and tryptophan in sub-tropical maize. Plant Sci 272:142–152
Shen R, FanJB CD, Chang W, Chen J, Doucet D, Yeakley J, Bibikova M, Wickham GE, McBride C, Steemers F, Garcia F, Kermani BG, Gunderson K, Oliphant A (2005) High-throughput SNP genotyping on universal bead arrays. Mutat Res 573:70–82. https://doi.org/10.1016/j.mrfmmm.2004.07.022
Shin JK, Know SJ, Lee JK, Min HK (2006) Genetic diversity of maize kernel starch-synthesis genes with SNPs. Genome 49:1287–1296
Shure M, Wessler S, Fedoroff N (1983) Molecular identification and isolation of the waxy locus in maize. Cell 35:225–233
Singh BD, Singh AK (2015) Marker-assisted plant breeding: principles and practices. Springer, New Delhi, India. https://doi.org/10.1007/978-81-322-2316-0
Stamp P, Eicke S, JampatongS, Le-Huy H, Jompuk C, EscherF, Streb S (2016) Southeast Asian waxy maize (Zea mays L.), a resource for amylopectin starch quality types? Plant Genet Res 1–8. https://doi.org/10.1017/S1479262116000101
Talukder ZA, Muthusamy V, Chhabra R et al. (2022) Combining higher accumulation of amylopectin, lysine and tryptophan in maize hybrids through genomics-assisted stacking of waxy1 and opaque2 genes. Sci Rep 12:706. https://doi.org/10.1038/s41598-021-04698-3
Tong J, Han Z, Han A, Liu X, Zhang S, Fu B, Zhu Y (2016) Sdt97, a point mutation in 5’ untranslated region, confers semi-dwarfism in rice. G3-Genes Genom Genet 6(6):1491–1502
Vrinten PL, Nakamura T (2000) Wheat granule-bound starch synthase I and II are encoded by separate genes that are expressed in different tissues. Plant Physiol 122(1):255–264. https://doi.org/10.1104/pp.122.1.255
Wessler S, Baran G, Varagona M, Dellaporta S (1986) Excision of Ds produces waxy proteins with a range of enzymatic activities. EMBO J 5:2427
Wu XY, Wu S, Long W, Chen D, Zhou G, Du J, Cai Q, Huang X (2019) New waxy allele wx-Reina found in Chinese waxy maize. Genet Resour Crop Evol 66:885–895
Xiaoyang W, Dan C, Yuqing L, Weihua L, XinmingY XL, Juan D, Lihui L (2017) Molecular characteristics of two new waxy mutations in China waxy maize. Mol Breed 37:27
Yang L, Wang M, We Wang, Yang W (2013) Marker-assisted selection for pyramiding the waxy and opaque-16 genes in maize using crosses and backcross schemes. Mol Breed 31:767–775. https://doi.org/10.1007/s11032-012-9830-8
Zhang WL, Yang WP, Wang MC, Wang W, Zeng GP, Chen ZW (2013) Increasing lysine content of waxy maize through introgression of opaque-2 and opaque-16 genes using molecular assisted and biochemical development. PLoS One 8:e56227. https://doi.org/10.1371/journal.pone.0056227
Zunjare RU, Hossain F, Muthusamy V, Baveja A, Chauhan HS, Bhat JS, Thirunavukkarasu N, Saha S, Gupta HS (2018) Development of biofortified maize hybrids through marker-assisted stacking of β-Carotene hydroxylase, Lycopene- ε-Cyclase and Opaque2 genes. Front Plant Sci 9:178
Acknowledgements
The authors sincerely acknowledge BHEARD and ICAR-Indian Agricultural Research Institute, New Delhi, for the financial support. The first author wishes to thank USAID, USA, for providing the BHEARD fellowship to undertake his Ph.D. program at ICAR-IARI. We thank IARI for providing the required lab and field facilities. We thank IIMR, Ludhiana, and AICRP (Maize) for providing the off-season nursery at Hyderabad. The authors sincerely thank Dr. B.M. Prasanna, CIMMYT, for providing the waxy source germplasm from where the waxy donor was developed. The help of CCSHAU, Uchani for sharing the parental inbreds is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
Genotyping of populations: ZAT, designing of markers and standardization of PCR protocol: RC, estimation of amylopectin: ZAT and RC, development of mapping populations: VM, statistical analysis: RUZ, manuscript writing: ZAT and FH, designing of the experiment: FH.
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Communicated by: Izabela Pawłowicz
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Talukder, Z.A., Chhabra, R., Muthusamy, V. et al. Development of novel gene-based markers for waxy1 gene and their validation for exploitation in molecular breeding for enhancement of amylopectin in maize. J Appl Genetics 64, 409–418 (2023). https://doi.org/10.1007/s13353-023-00762-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-023-00762-y