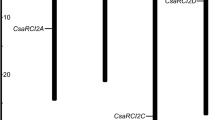
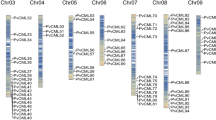
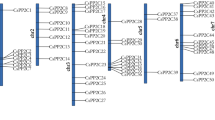
Abstract
MAP65 is a microtubule-binding protein family in plants and plays crucial roles in regulating cell growth and development, intercellular communication, and plant responses to various environmental stresses. However, MAP65s in Cucurbitaceae are still less understood. In this study, a total of 40 MAP65s were identified from six Cucurbitaceae species (Cucumis sativus L., Citrullus lanatus, Cucumis melo L., Cucurbita moschata, Lagenaria siceraria, and Benincasa hispida) and classified into five groups by phylogenetic analysis according to gene structures and conserved domains. A conserved domain (MAP65_ASE1) was found in all MAP65 proteins. In cucumber, we isolated six CsaMAP65s with different expression patterns in tissues including root, stem, leaf, female flower, male flower, and fruit. Subcellular localizations of CsaMAP65s verified that all CsaMAP65s were localized in microtubule and microfilament. Analyses of the promoter regions of CsaMAP65s have screened different cis-acting regulatory elements involved in growth and development and responses to hormone and stresses. In addition, CsaMAP65-5 in leaves was significantly upregulated by salt stress, and this promotion effect was higher in cucumber cultivars with salt tolerant than that without salt tolerant. CsaMAP65-1 in leaves was significantly upregulated by cold stress, and this promotion was higher in cold-tolerant cultivar than intolerant cultivar. With the genome-wide characterization and phylogenetic analysis of Cucurbitaceae MAP65s, and the expression profile of CsaMAP65s in cucumber, this study laid a foundation for further study on MAP65 functions in developmental processes and responses to abiotic stress in Cucurbitaceae species.










Similar content being viewed by others
Data availability
All data are presented in the main manuscript and the additional supplemental files.
References
Casas-Mollano JA, Lao NT, Kavanagh TA (2006) Intron-regulated expression of SUVH3, an Arabidopsis Su(var)3–9 homologue. J Exp Bot 57(12):3301–3311
Chan J, Rutten T, Lloyd C (1996) Isolation of microtubule-associated proteins from carrot cytoskeletons: A 120 kDa map decorates all four microtubule arrays and the nucleus. Plant J 10(2):251–259
Chang HY, Smertenko AP, Igarashi H, Dixon DP, Hussey PJ (2005) Dynamic interaction of NtMAP65-1a with microtubules in vivo. J Cell Sci 118(14):3195–3201
Chao J, Kong Y, Wang Q, Sun Y, Gong D, Lv J et al (2015) MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Yichuan 37(1):91–97
Ding D, Muthuswamy S, Meier I (2012) Functional interaction between the Arabidopsis orthologs of spindle assembly checkpoint proteins MAD1 and MAD2 and the nucleoporin NUA. Plant Mol Biol 79(3):203–216
Dorn A, Puchta H (2019) DNA Helicases as Safekeepers of genome stability in plants. Genes 10(12):1028
Duraisamy GS, Mishra AK, Kocabek T, Matousek J (2016) Identification and characterization of promoters and cis-regulator elements of genes involved in secondary metabolites production in hop (Humulus lupulus. L). Comput Biol Chem 64:346–352
Dvořák Tomaštíková E, Rutten T, Dvořák P, Tugai A, Ptošková K, Petrovská B et al (2020) Functional divergence of microtubule-associated TPX2 family members in Arabidopsis thaliana. Int J Mol Sci 21(6):2183
Fache V, Gaillard J, Van Damme D, Geelen D, Neumann E, Stoppin-Mellet V et al (2010) Arabidopsis kinetochore fiber-associated MAP65-4 cross-links microtubules and promotes microtubule bundle elongation. Plant Cell 22(11):3804–3815
Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A et al (2013) STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41(D1):D808-D815
Gao YF, Liu JK, Yang FM, Zhang GY, Wang D, Zhang L et al (2020) The WRKY transcription factor WRKY8 promotes resistance to pathogen infection and mediates drought and salt stress tolerance in Solanum lycopersicum. Physiol Plant 168(1):98–117
Garcia-Mas J, Benjak A, Sanseverino W, Bourgeois M, Mir G, González VM et al (2012) The genome of melon (Cucumis melo L.). Proc Natl Acad Sci U S Am 109(29):11872–11877
Guo LB, Ho CMK, Kong ZS, Lee YRJ, Qian Q, Liu B (2009) Evaluating the microtubule cytoskeleton and its interacting proteins in monocots by mining the rice genome. Ann Bot 103(3):387–402
Guo S, Zhang J, Sun H, Salse J, Lucas WJ, Zhang H et al (2013) The draft genome of watermelon (citrullus lanatus) and resequencing of 20 diverse accessions. Nat Genet 45(1):51–58
Guo YP, Shi JJ, Zhou MQ, Yu Y, Wang C (2020) Drought and salt tolerance analysis of BpbZIP1 gene in birch and ABRE element binding identification. For Res 33(5):68–76
Ho CM, Lee YR, Kiyama LD, Dinesh-Kumar SP, Liu B (2012) Arabidopsis microtubule-associated protein MAP65-3 cross-links antiparallel microtubules toward their plus ends in the phragmoplast via its distinct C-terminal microtubule binding domain. Plant Cell 24(5):2071–2085
Honghe S, Shan W, Guoyu Z, Chen J, Shaogui G et al (2018) Karyotype stability and unbiased fractionation in the paleo-allotetraploid cucurbita genomes. Mol Plant 10(10):1293–1306
Hornikova L, Brustikova K, Forstova J (2020) Microtubules in Polyomavirus Infection. Viruses-Basel 12(1):121
Hussey PJ, Hawkins TJ, Igarashi H, Kaloriti D, Smertenko A (2002) The plant cytoskeleton: recent advances in the study of the plant microtubule-associated proteins MAP-65, MAP-190 and the Xenopus MAP215-like protein, MOR1. Plant Mol Biol 50(6):915–924
Jiang CJ, Sonobe S (1993) Identification and preliminary characterization of a 65-kDa higher-plant microtubule-associated protein. J Cell Sci 105:891–901
Jiang Y, Liang G, Yu D (2012) Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol Plant 5(6):1375–1388
Kelley LA, Sternberg MJE (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4(3):363–371
Kollarova E, Forero AB, Stillerova L, Prerostova S, Cvrckova F (2020) Arabidopsis Class II formins AtFH13 and AtFH14 can form heterodimers but exhibit distinct patterns of cellular localization. Int J Mol Sci 21(1):348
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y et al (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327
Li Q, Li H, Huang W, Xu Y, Zhou Q, Wang S et al (2019) A chromosome-scale genome assembly of cucumber (cucumis sativus L.). GigaScience 8(6):giz072
Li H, Sun BJ, Sasabe M, Deng XG, Machida Y, Lin HH et al (2017) Arabidopsis MAP65-4 plays a role in phragmoplast microtubule organization and marks the cortical cell division site. New Phytol 215(1):187–201
Li H, Zeng X, Liu ZQ, Meng QT, Yuan M, Mao TL (2009) Arabidopsis microtubule-associated protein AtMAP65-2 acts as a microtubule stabilizer. Plant Mol Biol 69(3):313–324
Liu ZQ, Shi LP, Yang S, Qiu SS, Ma XL, Cai JS et al (2021) A conserved double-W box in the promoter of CaWRKY40 mediates autoregulation during response to pathogen attack and heat stress in pepper. Mol Plant Pathol 22(1):3–18
Lucas JR, Courtney S, Hassfurder M, Dhingra S, Bryant A, Shaw SL (2011) Microtubule-associated proteins MAP65-1 and MAP65-2 positively regulate axial cell growth in etiolated Arabidopsis hypocotyls. Plant Cell 23(5):1889–1903
Mao T, Jin L, Li H, Liu B, Yuan M (2005) Two microtubule-associated proteins of the Arabidopsis MAP65 family function differently on microtubules. Plant Physiol 138(2):654–662
Meng QT, Du JZ, Li JJ, Lu XM, Zeng XA, Yuan M et al (2010) Tobacco microtubule-associated protein, MAP65-1c, bundles and stabilizes microtubules. Plant Mol Biol 74(6):537–547
Mishra M, Huang JQ, Balasubramanian MK (2014) The yeast actin cytoskeleton. FEMS Microbiol Rev 38(2):213–227
Muller S, Smertenko A, Wagner V, Heinrich M, Hussey PJ, Hauser MT (2004) The plant microtubule-associated protein AtMAP65-3/PLE is essential for cytokinetic phragmoplast function. Curr Biol 14(5):412–417
Nishizawa-Yokoi A, Yamaguchi N (2018) Gene expression and transcription factor binding tests using mutated-promoter reporter lines. Methods Mol Biol (Clifton, N.J.) 1830:291–305
Parrotta L, Faleri C, Cresti M, Cai G (2017) Proteins immunologically related to MAP65-1 accumulate and localize differentially during bud development in Vitis vinifera L. Protoplasma 254(4):1591–1605
Portran D, Zoccoler M, Gaillard J, Stoppin-Mellet V, Neumann E, Arnal I et al (2013) MAP65/Ase1 promote microtubule flexibility. Mol Biol Cell 24(12):1964–1973
Rose AB, Elfersi T, Parra G, Korf I (2008) Promoter-proximal introns in Arabidopsis thaliana are enriched in dispersed signals that elevate gene expression. Plant Cell 20(3):543–551
Sasabe M, Machida Y (2006) MAP65: a bridge linking a MAP kinase to microtubule turnover. Curr Opin Plant Biol 9(6):563–570
Smertenko AP, Chang HY, Sonobe S, Fenyk SI, Weingartner M, Bogre L et al (2006) Control of the AtMAP65-1 interaction with microtubules through the cell cycle. J Cell Sci 119(15):3227–3237
Smertenko AP, Chang HY, Wagner V, Kaloriti D, Fenyk S, Sonobe S et al (2004) The Arabidopsis microtubule-associated protein AtMAP65-1: Molecular analysis of its microtubule bundling activity. Plant Cell 16(8):2035–2047
Soga K, Kotake T, Wakabayashi K, Hoson T (2012) Changes in the transcript levels of microtubule-associated protein MAP65-1 during reorientation of cortical microtubules in azuki bean epicotyls. Acta Physiol Plant 34(2):533–540
Stoppin-Mellet V, Fache V, Portran D, Martiel JL, Vantard M (2013) MAP65 coordinate microtubule growth during bundle formation. PLoS One 8(2):e56808
Struk S, Dhonukshe P (2014) MAPs: cellular navigators for microtubule array orientations in Arabidopsis. Plant Cell Rep 33(1):1–21
Tobias LM, Spokevicius AV, McFarlane HE, Bossinger G (2020) The cytoskeleton and its role in determining cellulose microfibril angle in secondary cell walls of woody tree species. Plants-Basel 9(1):90
Vavrdova T, Krenek P, Ovecka M, Samajova O, Flokova P, Illesova P et al (2020) Complementary superresolution visualization of composite plant microtubule organization and dynamics. Front Plant Sci 11:693
Wang C, Zhang LJ, Huang RD (2011) Cytoskeleton and plant salt stress tolerance. Plant Signal Behav 6(1):29–31
Wang XF, Mao TL (2019) Understanding the functions and mechanisms of plant cytoskeleton in response to environmental signals. Curr Opin Plant Biol 52:86–96
Wicker-Planquart C, Stoppin-Mellet V, Blanchoin L, Vantard M (2004) Interactions of tobacco microtubule-associated protein MAP65-1b with microtubules. Plant J 39(1):126–134
Wu S, Shamimuzzaman M, Sun H, Salse J, Sui X et al (2017) The bottle gourd genome provides insights into cucurbitaceae evolution and facilitates mapping of a papaya ring-spot virus resistance locus. Plant J 92(5):963–975
Xie D, Xu Y, Wang J, Liu W, Zhang Z (2019) The wax gourd genomes offer insights into the genetic diversity and ancestral cucurbit karyotype. Nat Commun 10(1):5158
Xu GX, Guo CC, Shan HY, Kong HZ (2012) Divergence of duplicate genes in exon-intron structure. Proc Natl Acad Sci USA 109(4):1187–1192
Xu R, Wang Y, Zheng H, Lu W, Wu C, Huang J et al (2015) Salt-induced transcription factor MYB74 is regulated by the RNA-directed DNA methylation pathway in Arabidopsis. J Exp Bot 66(19):5997–6008
Yang BJ, Wendrich JR, De Rybel B, Weijers D, Xue HW (2020) Rice microtubule-associated protein IQ67-DOMAIN14 regulates grain shape by modulating microtubule cytoskeleton dynamics. Plant Biotechnol J 18(5):1141–1152
Zhang HC, Deng XG, Sun BJ, Van SL, Kang ZS, Lin HH et al (2018) Role of the BUB3 protein in phragmoplast microtubule reorganization during cytokinesis. Nature Plants 4(7):485–494
Zhang Q, Lin F, Mao TL, Nie JN, Yan M, Yuan M et al (2012) Phosphatidic acid regulates microtubule organization by interacting with MAP65-1 in response to salt stress in Arabidopsis. Plant Cell 24(11):4555–4576
Zhang Y, Yu H, Yang X, Li Q, Ling J, Wang H et al (2016) CsWRKY46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner. Plant Physiol Biochem 108:478–487
Zhao W, Chen Z, Liu X, Che G, Zhang X (2017) Cslfy is required for shoot meristem maintenance via interaction with wuschel in cucumber (Cucumis sativus). New Phytol 218(1):344–356
Funding
This work was financially supported by the National Natural Science Foundation of China (No. 3200180213 and No. 31972478), the China Agriculture Research System (NO. CARS-23) and the Key Research and Development Program of Ningxia (2021BBF02005).
Author information
Authors and Affiliations
Contributions
S.M. and Y.Q.T conceived and designed the experiments; M.T.L, T.T.J, X.Y.W, and S.H.L performed the experiments and collected the data; M.T.L executed the data analyses; all authors contributed to the interpretation of the results; M.T.L, S.M., Y.Q.T, and L.H.G. wrote the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
No conflict of interest in this research work.
Additional information
Communicated by: Izabela Pawłowicz
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liang, M., Ji, T., Wang, X. et al. Comprehensive analyses of microtubule-associated protein MAP65 family genes in Cucurbitaceae and CsaMAP65s expression profiles in cucumber. J Appl Genetics 64, 393–408 (2023). https://doi.org/10.1007/s13353-023-00761-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-023-00761-z